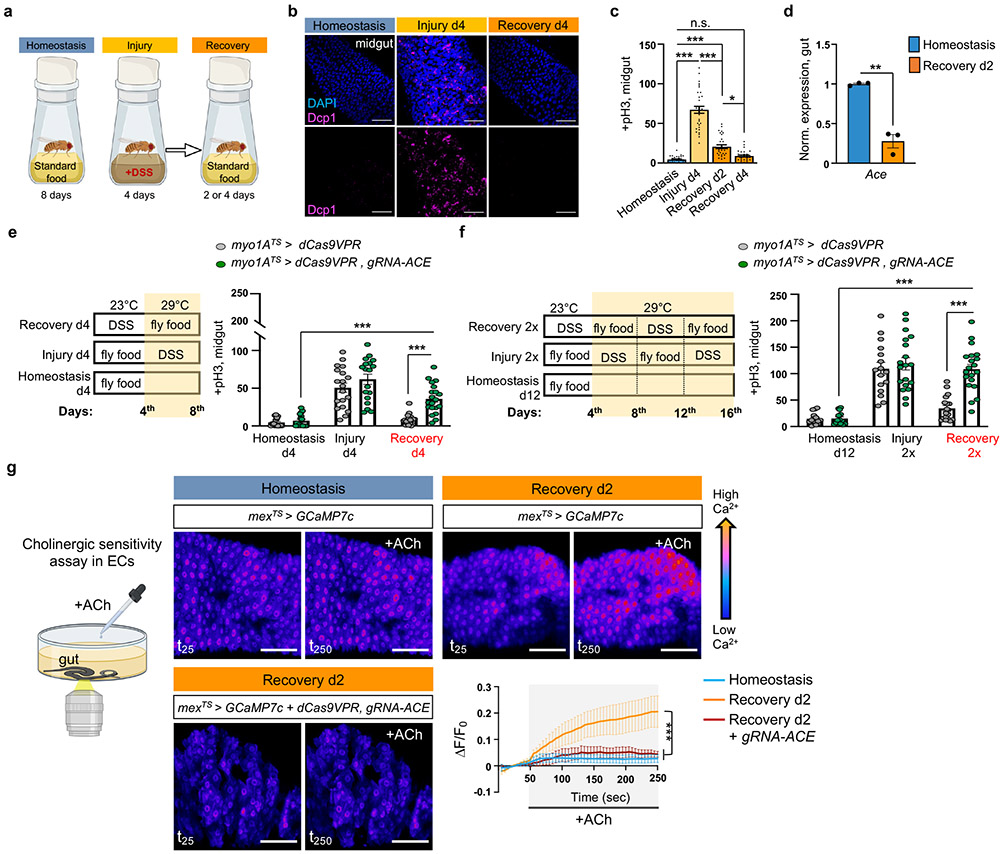
Fig.1 ∣. ACh sensitivity is required for recovery.
a, Experimental design illustration. Midgut of OreR flies with cell death marker anti-Dcp1 (Drosophila caspase 1, magenta) and DAPI (nuclei, blue). Conditions like 1a at 29°C. Images are representative of two independent experiments with similar results. scale bar: 50μm. c, Mitotic division counts of proliferating ISCs in the midgut of Ore R flies with anti-PhoshoHistone-3 (pH3) staining (conditions like Fig.1b). n=29 xDunn’s Kruskal-Wallis test : p=0.02 (Rec. d2 vs Rec. d4). Black dots indicate counts per gut. d, Expression levels of Ace in Ore R guts. Normalized to Homeostasis. n=3 biologically independent samples per condition (two-tailed t-test, p=0.0011). e-f, Experimental design and graphs of pH3+ counts from control (myo1ATS>dCas9VPR) and flies with Ace conditionally overexpressed in ECs (myo1ATS>dCas9VPR, gRNA-Ace). Conditional perturbations with temperature-sensitive Gal4 inhibitor, Tubulin-Gal80TS(TS), allow Gal4 expression only >27°C. control: n=18(Homeostasis),19(Injury), 20(Recovery), 15(Homeostasis d12), 16(Injury 2x), 18(Recovery 2x) guts ; gRNA-Ace: n=17(Homeostasis), 18(Injury), 20(Recovery),14(Homeostasis d12),19(Injury 2x), 20(Recovery 2x) guts, from 3 independent experiments. Sidak’s and Tukey’s two-way Anova. g, Assay illustration and color-coded sequential frames of midgut before (t25) and after (t250) ACh administration of flies conditionally expressing the Ca2+ reporter GCAMP7c with the EC-driver mex1-Gal4 (mexTS >GCaMP7c) and flies overexpressing Ace (mexTS >GCaMP7c+dCas9VPR,gRNA-Ace, Videos S1-S3). Recovery: 2 days in standard food (29°C) after 4 days of DSS-feeding (23°C). Homeostasis: conditions like Recovery without DSS-feeding. Accompanying graph: average relative fluorescence intensity (ΔF/F0) per frame (5 sec/frame) and genotype. n=7 (Homeostasis), n=7 (Recovery d2), n=5 (Recovery d2, mexTS> gRNA-Ace) guts, from 3 independent experiments. Tukey’s two-way Anova. Individual ΔF/F0 shown on Ext. Data Fig.2i. scale bar: 50μm *: 0.05>p>0.01, **: 0.01<p<0.001, ***: p<0.001, ns: non-significant. Data are presented as mean values ± SEM. Illustrations with Biorender.com