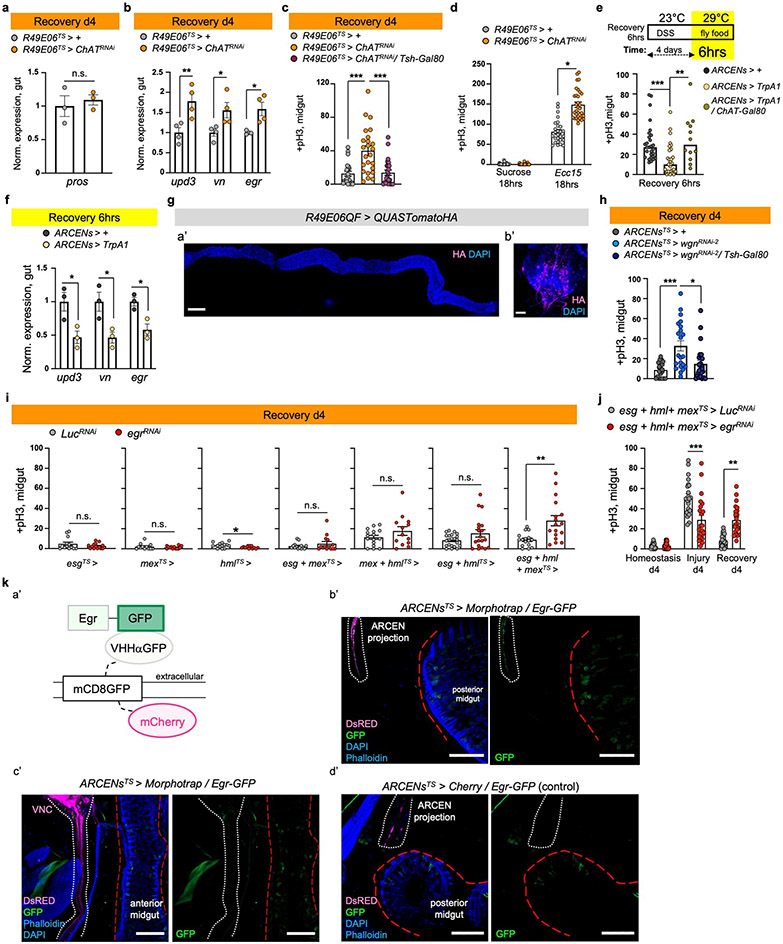
Ext. Data Fig.7 ∣. Egr-ARCENs signaling promotes recovery.
a-b, Pros, upd3, vn, egr expression levels from control (R49E06TS>+) guts and guts after knocking down ChAT in R49E06-neurons (R49E06TS>ChATRNAi). Conditions like Fig. 1e. a: n=3 biologically independent samples. b: n=4 biologically independent samples. Normalized to R49E06TS>+. Statistics: (a) two-tailed t-test, (b) Sidak’s two-way Anova: p=0.0038 (upd3), p=0.0412 (vn), p=0.0298(egr). c, pH3+ counts from R49E06TS>+ (control), R49E06TS>ChATRNAi flies and flies co-expressing the VNC repressor (R49E06TS>ChATRNAi/Tsh-Gal80). n= 21 (control), n=24 (R49E06TS>ChATRNAi, R49E06TS>ChATRNAi/Tsh-Gal80) guts examined over 3 independent experiments. Statistics: Dunn’s Kruskal-Wallis test: p=0.0004. Conditions like Fig. 1e. d, pH3+ counts from R49E06TS>+ and R49E06TS>ChATRNAi flies after oral Ecc15 infection or 5% sucrose feeding (like Ext. Data Fig.2g). n=27 (R49E06TS>+ per condition), n=24 (R49E06TS>ChATRNAi, sucrose), n=32 (R49E06TS>ChATRNAi, Ecc15) guts examined over 3 independent experiments. Statistics: Dunn’s Kruskal-Wallis test: p=0.013. e, Experimental schematic and pH3+ counts, from control (ARCENs>+) files, flies with 6hrs thermo-activation of ARCENs with theTrpA1 channel (ARCENs>TrpA1) and when cholinergic neurons are inhibited (ARCENs>TrpA1/ChAT-Gal80). n=28 (control), n=29 (ARCENs>TrpA1), n=12 (ARCENs>TrpA1/ChAT-Gal80) guts examined over 2 independent experiments. Dunn’s Kruskal-Wallis test: p= 0.0005(control vs TrpA1), p= 0.0069 (TrpA1 vs TrpA1/ChAT-Gal80). f, upd3, vn, egr expression levels from ARCENs>+ (control) and ARCENs>TrpA1 guts. Conditions like Ext. Data Fig.7e. n=3 biologically independent samples per genotype. Normalized to ARCENs>+. Sidak’s two-way Anova: p=0.0114 (upd3), p=0.0109 (vn), p=0.0461(egr). g, Validation of R49E06QF. a’: gut (scale bar: 200μm), b’: posterior VNC (scale bar 50μm). Images are representative of 2 independent experiments with similar results. h, pH3+ counts from control (ARCENsTS>+) flies, flies with TNF receptor wgn reduced in ARECNs (ARCENsTS>wgnRNAi-2) and flies co-expressing the VNC repressor (ARCENsTS>wgnRNAi-2/Tsh-Gal80). Conditions like Fig.1e. n=21 (control), n=22 (ARCENsTS>wgnRNAi-2), n=25(ARCENsTS>wgnRNAi-2/Tsh-Gal80) guts examined over 3 independent experiments. Dunn’s Kruskal-Wallis: p=0.0004(control vs wgnRNAi-2), p=0.0105(wgnRNAi-2 vs wgnRNAi-2 /Tsh-Gal80). i, pH3+ from control (LucRNAi) and flies with egr reduced (egrRNAi) in PCs (esgTS>), ECs (mexTS>) and hemocytes (hmlTS>). Conditions like Fig.1e. esgTS: n=13 (control), n=14 (egrRNAi) guts examined over 2 independent experiments (two-tailed Mann-Whitney test). mexTS: n=12 (control), n=13 (egrRNAi) guts examined over 2 independent experiments (two-tailed Mann-Whitney test). hmlTS: n=14 (control), n=13 (egrRNAi) guts examined over 2 independent experiments (two-tailed Mann-Whitney test, p=0.028). esg+mexTS: n=14 guts per genotype examined over 2 independent experiments (two-tailed Mann-Whitney test). mex+hmlTS: n=16 (control), n=13 (egrRNAi) guts examined over 2 independent experiments (two-tailed t-test). esg+hmlTS: n=21 (control), n=19 (egrRNAi) guts examined over 3 independent experiments (two-tailed Mann-Whitney test). esg+mex+hmlTS: n=16 (control), n=19 (egrRNAi) guts examined over 3 independent experiments (two-tailed Mann-Whitney test). j, pH3+ counts from esg+hml+mexTS>LucRNAi (control) and esg+hml+mexTS>egrRNAi flies. Conditions like Fig.1e. control: n=23 (Hom.), n=22 (Injury), n=40 (Rec.) guts; egrRNAi: n=24 (Hom.), n=22 (Injury), n=39 (Rec.) guts examined over 3 independent experiments. Statistics: Sidak’s two-way Anova. k, a’: Schematic of Egr-GFP bound to extracellular morphotrap (VHHaGFP). b’-d’: Images from the abdomen (b’,d’) and thorax (c’) of flies expressing the morhotrap in ARCENs while expressing Egr-GFP (ARCENsTS>morphotrap/Egr-GFP) and of control flies without the morphotrap (ARCENsTS>Cherry/Egr-GFP). white dotted lines: ARCEN projections. red lines: midgut (b’,d’: posterior, c’: anterior). Sectioning like Fig.4a. anti-DsRed: Morphotrap and Cherry (magenta). Anti-GFP: Egr (green), Phalloidin: muscle (blue), DAPI: nuclei (blue). Images are representative of 2 independent experiments with similar results. scale bar 50μm. n.s.: non-significant ,*: 0.05>p>0.01, **: 0.01<p<0.001, ***: p<0.001. Data are presented as mean values ± SEM.