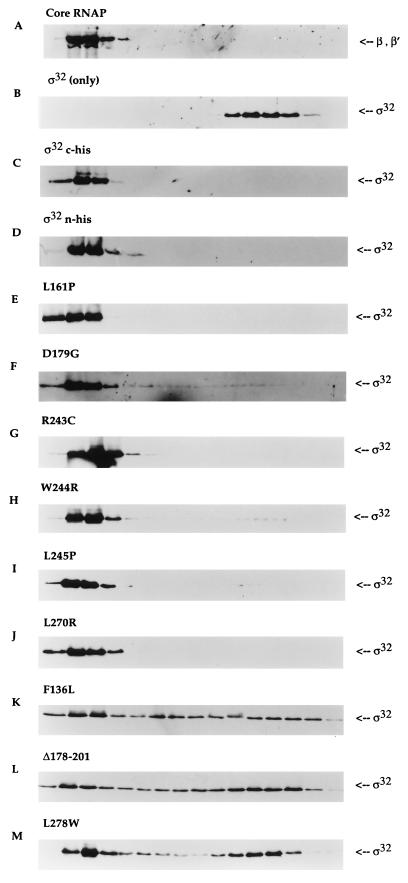
FIG. 3.
Core RNAP binding analysis of ς32 proteins by glycerol gradient sedimentation. Equimolar concentrations of core RNAP and different ς32 proteins (final concentration of 100 nM) were allowed to reconstitute at 30°C for 15 min in a buffer containing 50 mM HEPES (pH 7.9) at 4°C, 0.1 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, and 10 mM MgCl2. The mixture was loaded onto the top of a 5-ml 15 to 35% glycerol gradient and centrifuged at 48,000 rpm for 24 h at 4°C in a Beckman SW50.1 rotor. Sixteen fractions were collected from the bottom of the tube and subjected to immunoblot analysis. The sedimentation patterns of each ς32 protein were detected with anti-ς32 serum. The positions of β and β′ subunits were determined with polyclonal antibodies to core RNAP. The leftmost region of each panel represents the bottom of the tube (35% glycerol). The positions of core RNAP were indistinguishable with (A) or without (data not shown) ς32. The sedimentation patterns are shown for ς32 only (B), for two different wild-type proteins with core RNAP (C and D), and (as indicated above each panel) for the mutant proteins with core RNAP (E through M).