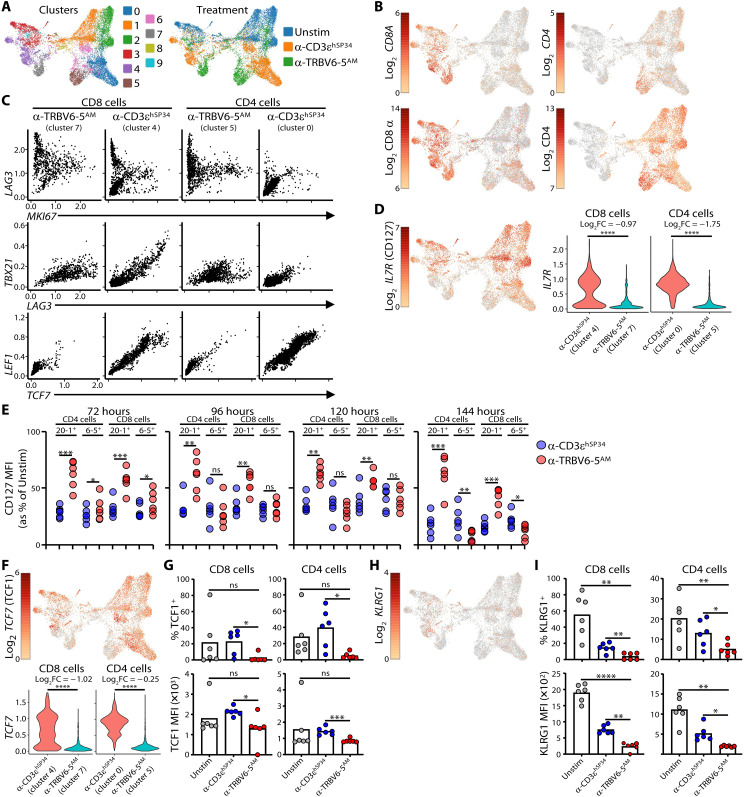
Fig. 6. Single-cell transcriptome analysis α-TRBV–stimulated cells.
(A and B) Purified T cells were cultured for 7 days with the indicated plate-bound antibodies or rested overnight without cytokines (Unstim) and then processed for single-cell RNA sequencing analysis. UMAP representation of clusters and treatment groups are shown in (A); CD4 and CD8 cells are identified in (B) using gene expression (top row) and barcoded antibodies (bottom row). (C) Imputed counts of the indicated genes were used in scatterplots to visualize coexpression patterns across clusters and cell types. (D) UMAP representation of IL7R expression (left) and violin plots (imputed expression, right). Log2FC, log2 fold change; ****P < 0.0001 [Wilcoxon rank sum test with Bonferroni correction, from the differentially expressed gene (DEG) analysis]. (E) Purified T cells were cultured without (Unstim) or with the indicated plate-bound antibodies and cell-surface expression of CD127 on the indicated subsets was monitored by flow cytometry at the indicated time points (n = 6 donors). *P < 0.05; **P < 0.01; ***P < 0.001 (paired t test). (F) UMAP representation of TCF7 expression (top row) and violin plots (imputed expression, bottom row). ****P < 0.0001 (Wilcoxon rank sum test with Bonferroni correction, from the DEG analysis). (G) Purified T cells were cultured for 7 days without (Unstim) or with the indicated plate-bound antibodies and intracellular TCF1 expression was measured in CD4 or CD8 cells. (H) UMAP representation of KLRG1 expression. (I) Purified T cells were cultured for 7 days without (Unstim) or with the indicated plate-bound antibodies and cell surface KLRG1 expression was measured on CD4 or CD8 cells. Data in (G) and (I) are the mean (bars) of percentage positive cells (top row) and MFI of positive cells (bottom row) for n = 6 donors. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (paired ANOVA with Dunnett’s method, comparing α-TRBV6-5AM to every other condition individually).