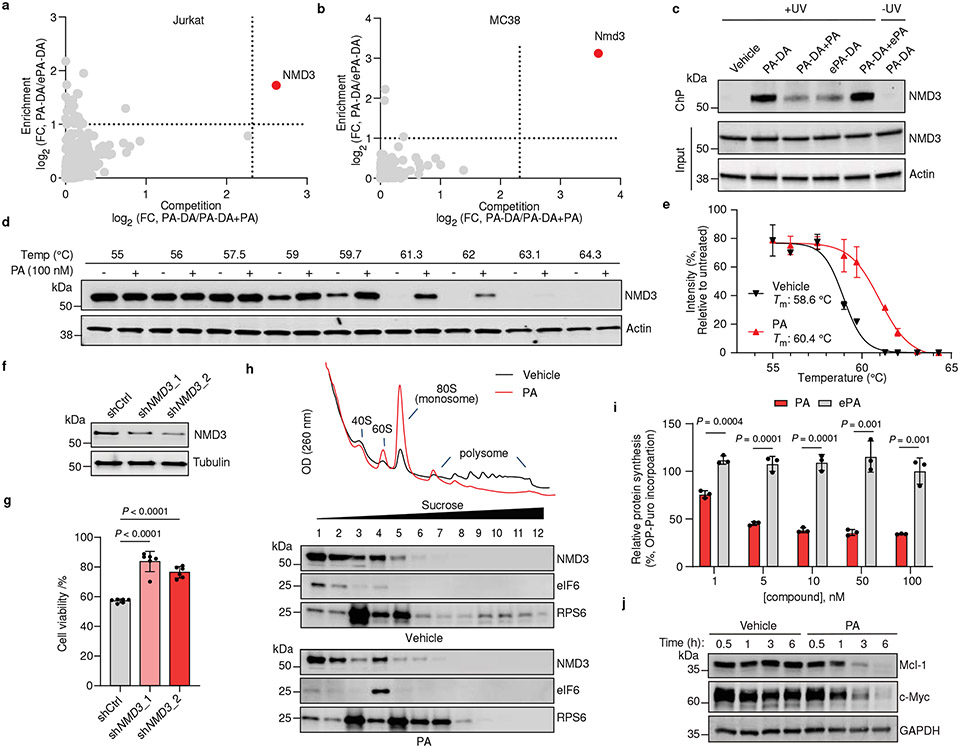
Fig. 4. Portimine A targets NMD3, prevents polysome formation and inhibits protein translation.
Chemoproteomic profiling of PA in (a) Jurkat and (b) MC38 cells. X-axes correspond to PA-DA (500 nM) enriched proteins competed by PA (8 ×); y-axes correspond to proteins enriched by PA-DA over ePA-DA (500 nM). Designated PA-specific targets in red (competed by active competitor > 5-fold; enriched by PA-DA > 2-fold; and > 4-fold competition difference between PA and ePA). Dotted lines indicate competition (x-axis) and enrichment (y-axis) thresholds. Data presented as mean of biological replicates (n = 2). See Supplementary Tables 7-9 for source data and Methods for additional details. c, Immunoblot of NMD3 engagement by PA-DA (500 nM) co-treated with PA or ePA (8 ×) as well as ePA-DA (500 nM). Representative of three biologically independent samples. d, Immunoblot of NMD3 thermal aggregation (CETSA) in the presence of PA (100 nM). Results representative of two biological replicates. e, Quantitation of the thermal aggregation curves (n = 2 biological replicates; mean ± s.d.). f, Immunoblot of NMD3 in shNMD3 Jurkat cells. g, PA (1 nM, 24 h) has reduced viability effects in Jurkat cells transduced with shRNA targeting NMD3 (n = 6 biological replicates; mean ± s.d.). Statistical analysis performed using unpaired two-tailed Student t-test; P-values are shown. h, Polysome profiling of Jurkat cells treated with PA (50 nM, 6 h). Results representative of three independent experiments. i, Protein synthesis in Jurkat cells inhibited by PA, but not ePA (6 h), as measured by O-propargyl puromycin (OPP) incorporation. Data normalized to vehicle and presented as mean ± s.d. across biological replicates (n = 3). Statistical analysis performed using multiple unpaired two-tailed Student t-test; P-values are shown. j, Immunoblot of Mcl-1 and c-Myc in Jurkat cells treated with PA (10 nM). Results representative of three biological replicates. For uncropped western blot images, see Supplementary Fig. 2.