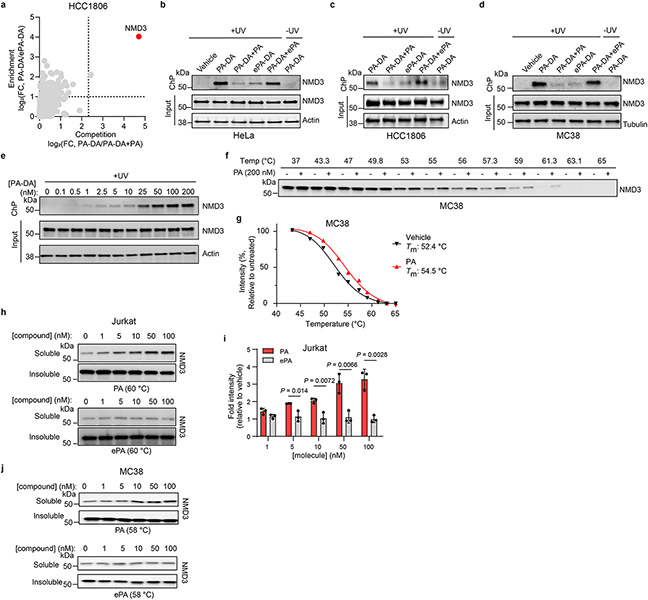
Extended Data Fig. 5 ∣. Chemical proteomic analysis reveals NMD3 is the target of portimine A.
(a) Chemical proteomic profiling with PA-DA in HCC1806. X-axis corresponds to PA-DA (500 nM) enriched proteins competed by PA (PA, 8 ×); y-axis corresponds to proteins enriched by PA-DA over ePA-DA (500 nM). Designated PA-specific targets in red (competed by active competitor > 5-fold; enriched by PA-DA > 2-fold; and > 4-fold competition difference between PA and ePA). Dotted lines indicate competition (x-axis) and enrichment (y-axis) thresholds. Data presented as mean of biological replicates (n = 2). See Supplementary Tables 7-9 for source data. (b-d) Immunoblot of NMD3 engagement by PA-DA (500 nM) co-treated with PA or ePA (8 ×) as well as by ePA-DA (500 nM) in HeLa (b), HCC1806 (c), and MC38 cells (d). (n = 2 biologically independent samples). (e) NMD3 is engaged by PA-DA in a dose-dependent manner in Jurkat cells. Results are representative of three independent experiments. (f-i) CETSA validation of NMD3 as a target of PA in Jurkat and MC38 cells. (f-g) Immunoblotting and quantitation of NMD3 thermal aggregation curves (mean ± s.d.) in MC38 cells treated with PA. (n = 3 biologically independent samples). (h-i) Dose-response (ITDR) fingerprint of NMD3 stabilization by PA in Jurkat cells and corresponding quantitation. Data presented as mean ± s.d. of biological replicated experiments (n = 3). Statistical analysis performed using multiple unpaired two-tailed Student t-test; P-values are shown. (j) Isothermal dose-response (ITDR) fingerprint of NMD3 stabilization by PA in MC38 cells (n = 3 biologically independent samples). For uncropped western blot images, see Supplementary Fig. 3.