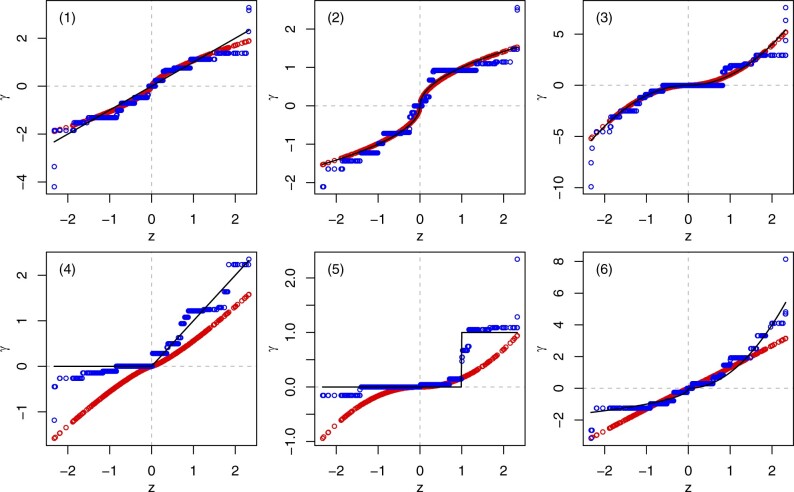
Figure 1.
Final prior effects (y-axis) against initial prior effects z (x-axis), under exponential calibration (red points on continuous curve) and isotonic calibration (blue points on discontinuous curve). The thin black line corresponds to perfect calibration (). We simulated the feature matrix X from a standard Gaussian distribution (n = 200, p = 500) and the initial prior effects z from a trimmed standard Gaussian distribution (trimmed below the 1% and above the 99% quantile). We set the true coefficients to (1), (2), (3), (4), (5), or (6). And we simulated the response vector y from Gaussian distributions with the means and the variance , where . While exponential calibration performs slightly better in the first three scenarios (top), isotonic calibration performs much better in the last three scenarios (bottom).