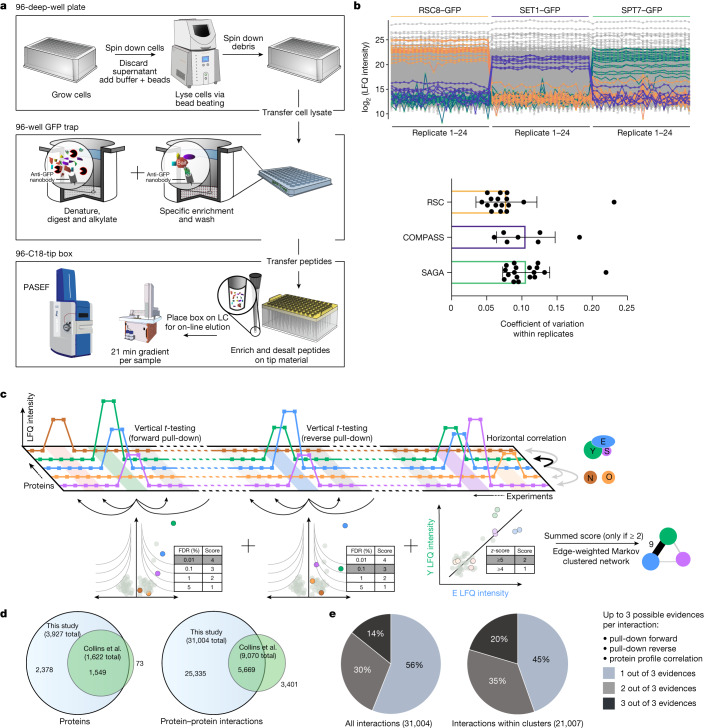
Fig. 1. A comprehensive and scalable interactomics technology.
a, Sample preparation in 96-well format and mass spectrometric measurement. Each strain of the GFP-tagged library is lysed by mechanical disruption and transferred to anti-GFP nanobody coated microtitre plates, where weak interactions are preserved by gentle washing. After enzymatic in-well digestion, resulting peptides are transferred to standardized C18-StageTips from which they are eluted directly into a standardized 60 samples per day gradient. Data are acquired in the PASEF scan mode on a trapped ion mobility—time-of-flight mass spectrometer. LC, liquid chromatography. b, Streamlined workflow and reduced transfer steps reduce the risk of manual errors and sample variation. Demonstration of workflow reproducibility and sensitivity on three nuclear complexes in biological replicates. A tagged member (bait) of each complex pulls down the known preys in very similar amounts based on label-free quantification (LFQ) intensities. Bottom, coefficient of variation (within 24 replicates), mean with standard deviation (n = 16 (RSC), 7 (COMPASS) and 18 (SAGA) complex members). c, Two-dimensional interaction scoring. Columns represent pull-down experiments in replicates (light colour). Squares depict intensities of detected proteins across the pull-down experiments. Three levels of evidence support each interaction: t-test of forward pull-down against complementary experiments, t-test of reverse pull-down, and protein profile correlation—the correlated abundance profile against all other proteins across all experiments (z-scored; Methods, ‘Protein correlation’). d, Overlap of proteins with at least one interactor and interactions detected in this study with the previous state-of-the-art network2. e, The proportion of interactions backed by multiple layers of evidence in the complete network and the network excluding inter-cluster interactions.