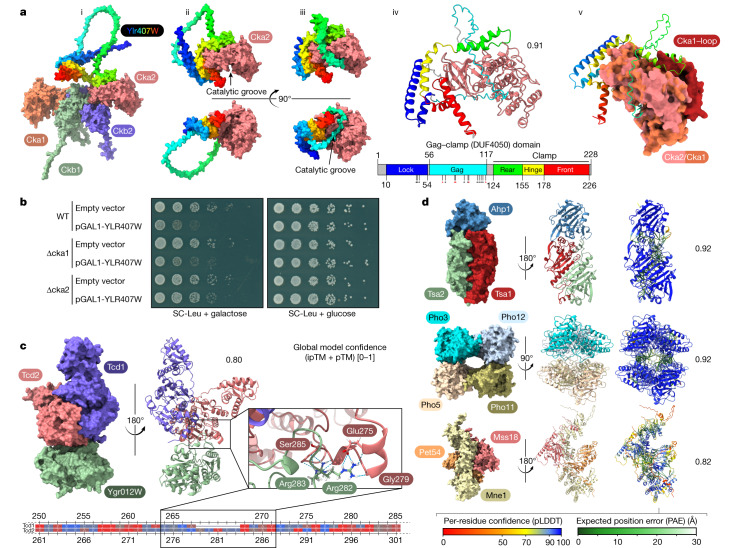
Fig. 5. Interaction-based structure prediction.
a, Structure and binding mode of YLR407Wp to Cka2 as modelled in AlphaFold-Multimer. (i) Competitive binding mode of the YLR407Wp front clamp binding domain with the CK2 holoenzyme. (ii) and (iii) Two major predicted models with the unstructured loop acting either as a lasso or a catalytic groove gag. (iv) Motifs of the conserved DUF4050 domain are coloured and named based on the modelled structure. Annotated phosphorylation sites are indicated with asterisks (black, reported twice in UniProt; red, reported three or more times). (v) Superposition of Cka1 and Cka2, with the clash of the Cka1-specific insertion loop with the rear-clamp domain highlighted. b, Serial dilution spotting of yeast strains carrying a Leu marker plasmid overexpressing YLR407W under the galactose inducible promoter or a negative control (empty vector). The growth defect phenotype caused by YLR407W overexpression is dependent on the presence of the interactor Cka2 but not on the non-interactor Cka1. c, Suggested binding mode of the two homologous proteins Tcd1 and Tcd2 with YGR012Wp. The alignment is coloured based on amino acid side chain hydrophobicity, with the highest in red to the lowest in blue. pTM, predicted template modeling score; ipTM, interface pTM. d, Selection of high-confidence complexes and their scoring (for an extended version see Supplementary Fig. 4).