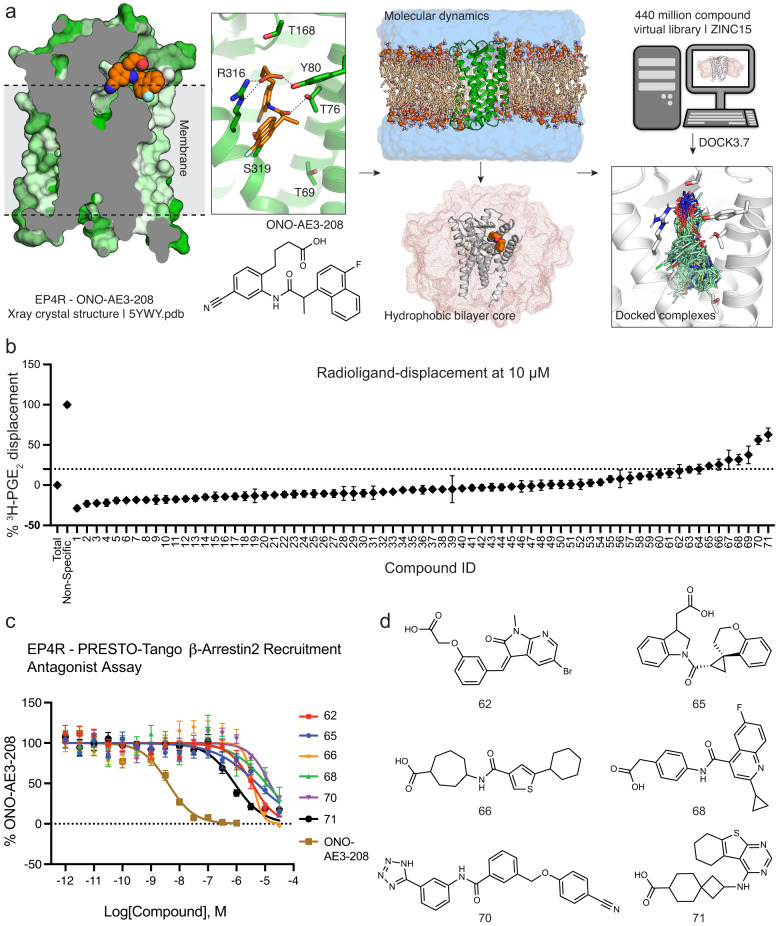
Fig. 1. Computational docking and in vitro testing for EP4R antagonists.
a The crystal structure of the EP4R-ONO-AE3-208 complex (PDB 5YWY)29 was targeted for docking. To reflect the low dielectric of the hydrophobic lipids surrounding the receptor, we equilibrated a POPC lipid bilayer around the protein using molecular dynamics simulations. About 440 million molecules were then docked against the EP4R structure. b Displacement of radiolabeled 3H-PGE2 by docking hits at 10 µM. c Concentration-response curves of docking hits using a PRESTO-Tango β-arrestin2 recruitment assay. d Docking hits demonstrating antagonist activity at EP4R. Data in b) represents mean ± SEM of three (for compounds from the ZINC15 library) or four (for compounds from the additional anion library) technical repetitions (see Supplementary Data 1). Data in c) represents mean ± SEM of three independent experiments. Source data are provided as a Source Data file.