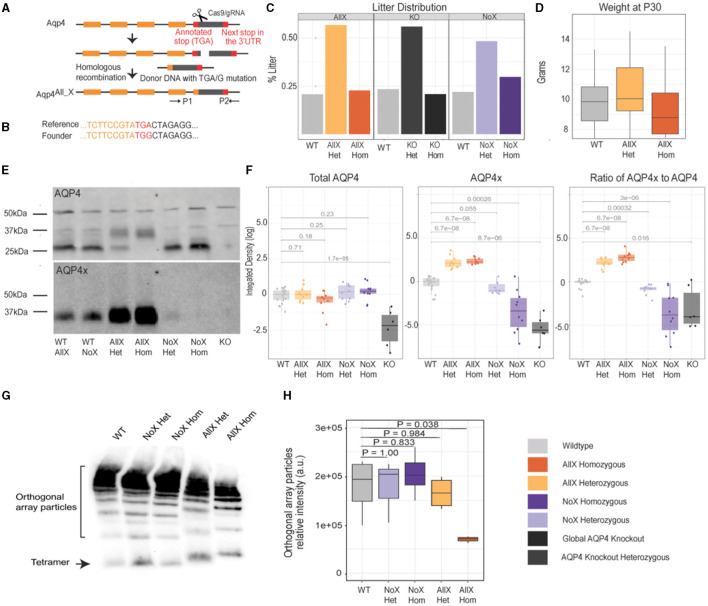
Figure 1.
Generation of an obligate AQP4 readthrough line and characterization of AQP4x production. (A) CRISPR/Cas9 construction of the AllX mutant mouse line. (B) Reference and founder sequence showing conversion of the TGA stop codon to a TGG (encoding tryptophan). (C) Litter distribution among the genotypes follows the Mendelian ratio. n (litters) = 13 Aqp4 (KO) line, 36 AllX line, and 32 NoX line. (D) Weight distribution at P30 between AllX and WT mice, n = 10 WT, 9 AllXHom, 22 AllXHet. (E) Western blot showing AQP4 and AQP4x expression, n = 3 per genotype, 1 global AQP4 KO mouse as a specificity control for the antibody. Total brain lysates probed with anti-AQP4 and anti-AQP4x antibodies show a significant increase in AQP4x compared to AQP4 in the AllX variants. AQP4 expression is enhanced in the no-readthrough variants and decreases in the AllX variants. The anti-AQP4 antibody exhibits two non-specific bands at 40 and 50 kDa, as evidenced by their presence in the global AQP4 knockouts (asterisk). (F) Quantification of Western blot as box plots (log10 scale) of AQP4 and AQP4x total and the relative ratio of AQP4x to AQP4 quantifying Western blot using ImageJ, normalized to 50 kDa band and wildtype. WT AllX and WT NoX merged and analyzed using the Wilcox test. (G) BN-PAGE with anti-AQP4 antibody showing orthogonal array particles and tetramer expression. (H) Quantification of orthogonal array particle signal from (G). n = 3–4 mice per genotype. One-way ANOVA, with post-hoc P-values from Dunnett's test.