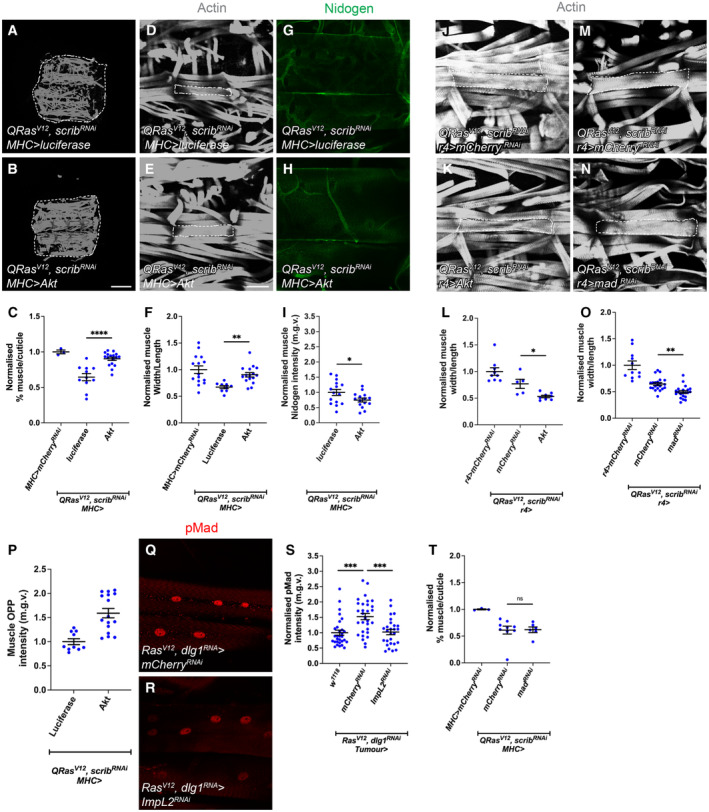
Figure EV3. Muscle overexpression of Akt (but not fat body Akt or mad RNAi overexpression) rescues atrophy in cachectic animals, and TGF‐ß signalling in the muscle is responsive to modulation in tumour insulin signalling but is not required for muscle integrity in cachexia.
-
A, BMuscle fillets stained with phalloidin (Actin) where luciferase or Akt was expressed under the control of MHC‐GAL4 in QRas V12 scrib RNAi tumour‐bearing animals.
-
CQuantification of normalised muscle detachment in (A, B). Wilt type control (MHC>mCherry RNAi , n = 3), UAS‐luciferase (n = 10), Akt (n = 17).
-
D, EMuscle segment (outlined) where luciferase or Akt was expressed under the control of MHC‐GAL4 in QRas V12 scrib RNAi tumour‐bearing animals.
-
FQuantification of normalised muscle width/length in (D, E). Wild type control (n = 15), luciferase (n = 9), Akt (n = 16).
-
G, HMuscle Nidogen staining where luciferase or Akt was expressed under the control of MHC‐GAL4 in QRas V12 scrib RNAi tumour‐bearing animals.
-
IQuantification of normalised muscle Nidogen staining in (G, H). luciferase (n = 16), Akt (n = 15).
-
J, KMuscle segment from QRas V12 scrib RNAi tumour‐bearing animals, where mCherry RNAi or Akt was expressed in the fat body (r4‐GAL4).
-
LQuantification of normalised muscle width/length in (J, K). r4>mCherry RNAi (n = 9), QRas V12 scrib RNAi ;r4>mCherry RNAi (n = 5), QRas V12 scrib RNAi ;r4>Akt (n = 8).
-
M, NMuscle segment from QRas V12 scrib RNAi tumour‐bearing animals, where mCherry RNAi or mad RNAi was expressed in the fat body (r4‐GAL4).
-
OQuantification of normalised muscle width/length in (M, N). r4>mCherry RNAi (n = 11), QRas V12 scrib RNAi ; r4>mCherry RNAi (n = 18), QRas V12 scrib RNAi ; r4>mad RNAi (n = 15).
-
PQuantification of normalised muscle OPP intensity where luciferase (n = 10) or Akt (n = 16) was expressed under the control of MHC‐GAL4 in QRas V12 scrib RNAi tumour‐bearing animals.
-
Q, RMuscle from Ras V12 dlg1 RNAi tumour‐bearing animals expressing either mCherry RNAi or ImpL2 RNAi in the tumour, where TGF‐ß signalling activation is indicated by pMad staining.
-
SQuantification of normalised muscle pMad intensity in (Q, R). w 1118 (n = 30), mCherry RNAi (n = 30), ImpL2 RNAi (n = 26).
-
TQuantification of normalised muscle detachment where MHC>mCherry RNAi (n = 5) or mCherry RNAi (n = 9) or lacZ RNAi ; mad RNAi (n = 6) was expressed under the control of MHC‐GAL4 in QRas V12 scrib RNAi tumour‐bearing animals.
Data information: Scale bar is 500 μm for muscle fillets (A, B), 100 μm for muscle segment atrophy measurements (D, E, J, K, M, N) and 50 μm for muscle Nidogen and pMad staining (G, H, Q, R). Muscles in tumour bearing animals were dissected at day 6 after tumour induction. Graphs are represented as Mean ± SEM, n = the number of samples. (*) P < 0.05 (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, (ns) P > 0.05. For experiments with two genotypes, two‐tailed unpaired student's t‐tests were used to test for significant differences. The Welch's correction was applied in cases of unequal variances. For experiments with more than two genotypes, significant differences between specific genotypes were tested using a one‐way ANOVA and a subsequent Šidák post‐hoc test.
