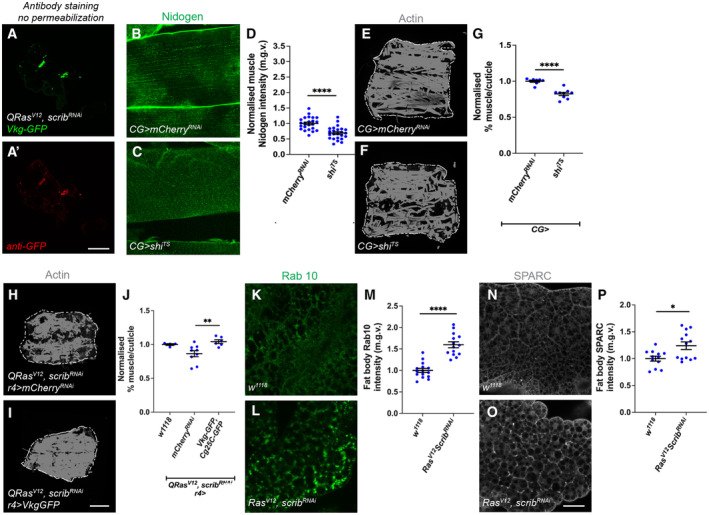
Figure EV4. Inhibition of fat body endocytosis causes muscle detachment, increasing Vkg expression in the fat body improves muscle integrity in tumour‐bearing animals.
-
A, A'Antibody staining against GFP without permeabilisation can detect Vkg‐GFP.
-
B, CNidogen staining in muscles of animals where mCherry RNAi or shi TS was expressed in the fat body via CG‐GAL4.
-
DQuantification of muscle nidogen staining in (B, C). mCherry RNAi (n = 23), shi TS (n = 24).
-
E, FMuscle fillets stained with phalloidin (Actin) from animals where mCherry RNAi or shi TS was expressed in the fat body via CG‐GAL4.
-
GQuantification of normalised muscle detachment in (E, F). mCherry RNAi (n = 8), shi TS (n = 9).
-
H, IMuscle fillets stained with phalloidin (Actin) from QRas V12 scrib RNAi tumour‐bearing animals, where mCherry RNAi or UAS‐VkgGFP;UAS‐Cg25C‐GFP was expressed in the fat body (r4‐GAL4). Day 6 muscles used here.
-
JQuantification of normalised muscle detachment in (H, I). w 1118 (n = 4), QRas V12 scrib RNAi ;r4>mCherry RNAi (n = 9), QRas V12 scrib RNAi ;r4>UAS‐VkgGFP;UAS‐Cg25C‐GFP (n = 6).
-
K, LFat body staining for Rab10 in w 1118 and QRas V12 scrib RNAi tumour‐bearing animals.
-
MQuantification of normalised Rab10 levels in (K, L). w 1118 (n = 15), QRas V12 scrib RNAi (n = 14).
-
N, OFat body staining for SPARC in w 1118 and QRas V12 scrib RNAi tumour‐bearing animals.
-
PQuantification of normalised SPARC levels in (N, O). w 1118 (n = 12), QRas V12 scrib RNAi (n = 13).
Data information: Scale bar is 500 μm for muscle fillets (E, F, H, I) 50 μm for muscle Nidogen and GFP staining (A, A′, B, C) and 50 μm for fat body staining (K, L, N, O). Muscles in Shi TS animals were dissected at day 5 ALH, muscles and fat body in tumour bearing animals were dissected at day 6 after tumour induction. Graphs are represented as Mean ± SEM, n = the number of samples. (*) P < 0.05 (**) P < 0.01, (****) P < 0.0001. For experiments with two genotypes, two‐tailed unpaired student's t‐tests were used to test for significant differences. The Welch's correction was applied in cases of unequal variances. For experiments with more than two genotypes, significant differences between specific genotypes were tested using a one‐way ANOVA and a subsequent Šidák post‐hoc test.
