Figure 1. An siRNA mini‐screen identifies CCR4‐NOT components as regulators of HCMV replication.
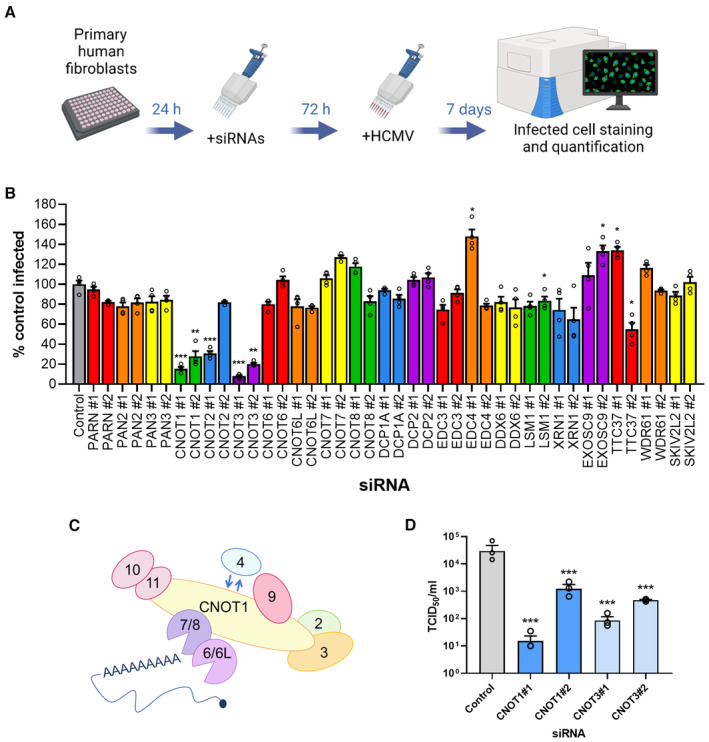
- Experimental set up for siRNA screen. Normal human dermal fibroblasts (NHDFs) were transfected at 20 nM with two independent siRNAs targeting each of 20 genes in 96‐well plates. Cells were subsequently infected with HCMV (AD169) at low MOI (0.05) and cells fixed 7 days post infection. Cells were identified by DAPI nuclear staining, and HCMV‐positive cells were identified by immunostaining for immediate early proteins and quantified in each entire well using high content imaging (Thermofisher CellInsight CX7 LZR).
- Screen results showing mean % infected cells (±SEM) of four biological experiments with technical duplicates, normalized to control siRNA treated cells. Statistical significance was established by ANOVA test with Dunnett multiple comparison correction compared with control siRNA, (*) P < 0.033, (**) P < 0.002, (***) P < 0.001, no asterisk: not significant.
- Schematic of human CCR4‐NOT complex.
- The impact of CNOT1 and CNOT3 depletion on released infectious viral titer was determined by replicating experimental conditions in (B) in 12‐well plates and establishing TCID50 from culture supernatants on NHDF cells, plotted as the mean ± SEM (n = 3 biological replicates). Statistical significance established by ANOVA test with Dunnett multiple comparison correction compared with control siRNA, (***) P < 0.001.
Source data are available online for this figure.
