Figure 2. CNOT1 and CNOT3 promote viral gene expression late in HCMV infection.
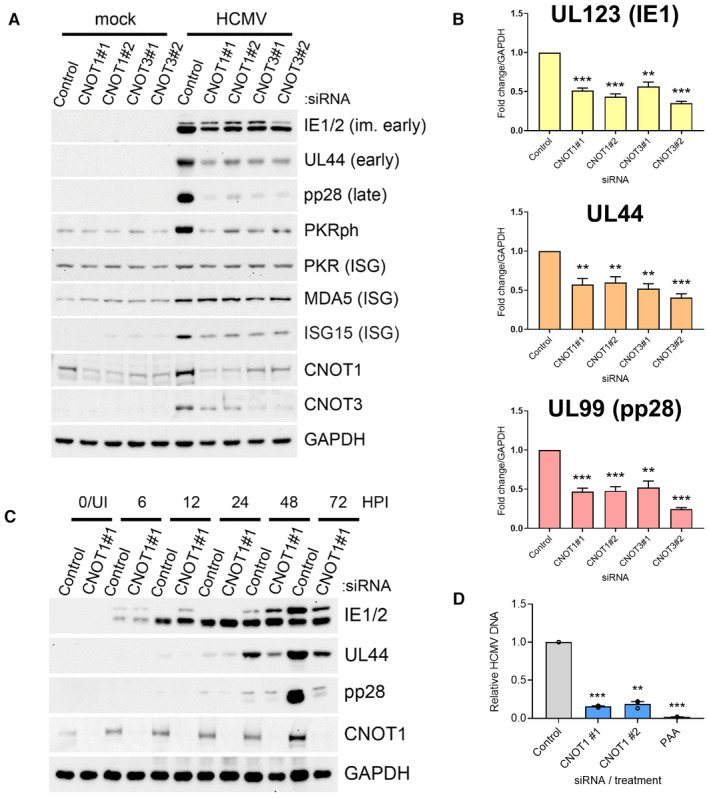
- Immunoblot analysis of lysates from NHDF cells transfected with the indicated siRNAs and infected with HCMV AD169 at MOI = 3, collected at 72 HPI. Lysates were probed for representative viral proteins from each temporal expression class, select ISGs (MDA5, ISG15, PKR) and activated T446‐phosphorylated PKR (PKRph) and GAPDH.
- RT‐qPCR analysis for viral mRNAs from RNA isolated from cells treated and infected as in (A). Protein names, where different from gene names, indicated in brackets. Mean ± SEM (n = 6 biological replicates) is plotted with statistical significance established by ANOVA test with Dunnett multiple comparison correction compared with control siRNA; (**) P < 0.002, (***) P < 0.001.
- Immunoblot analysis of lysates from NHDF cells transfected with control or CNOT1 siRNAs and infected with HCMV AD169 at MOI = 3 collected at the indicated times post infection. Lysates were probed for representative viral proteins indicated, CNOT1 to confirm knockdown and a cellular loading control (GAPDH).
- NHDFs transfected with control or CNOT1‐specific siRNAs were infected with HCMV AD169 at MOI = 3, as in (A). At 72 hpi, total DNA was collected and HCMV DNA abundance was quantified by qPCR. Untransfected cells were treated in parallel with viral DNA polymerase inhibitor PAA after virus inoculation as a control. Mean ± SEM (n = 3 biological replicates) is plotted with statistical significance established by ANOVA test with Dunnett multiple comparison correction compared with control siRNA; (**) P < 0.002, (***) P < 0.001.
Source data are available online for this figure.
