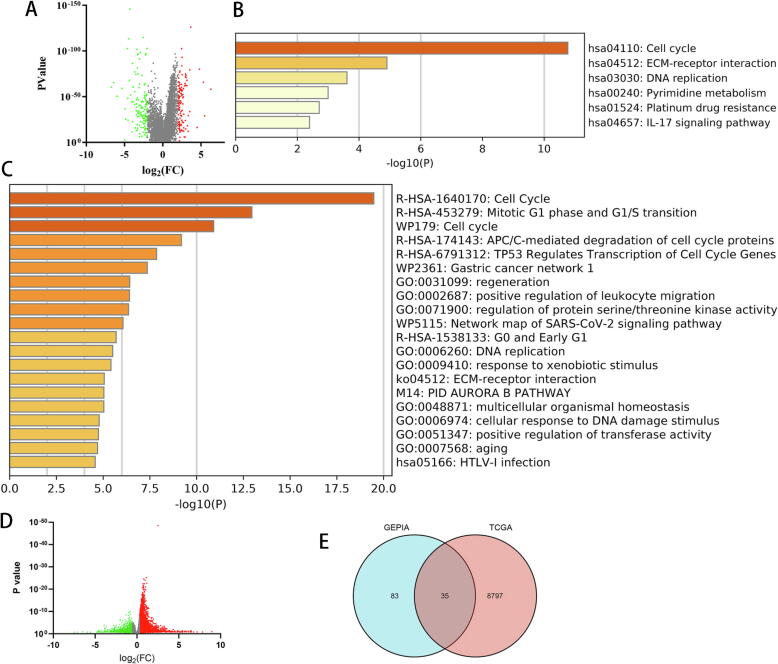
Fig. 1.
Identification of differentially expressed genes (DEGs) in LIHC. (A) Volcano plot of DEGs between LIHC tissues and normal liver tissues using GEPIA. KEGG (B) and GO (C) functional enrichment analysis of DEGs in LIHC versus adjacent tissues. (D) Volcano plot of high-MYC and low-MYC LIHC groups using datasets from TCGA. (E) Venn diagram of overlapping DEGs associated with high MYC expression and LIHC-specific expression.