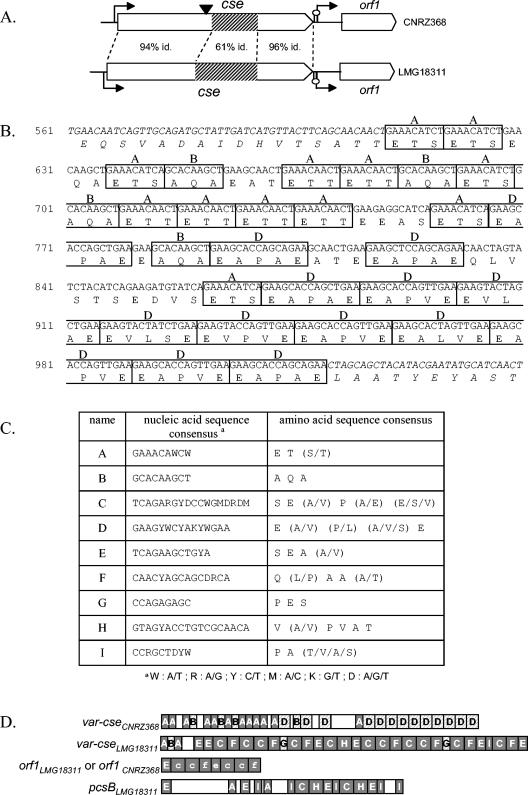
FIG. 2.
var-cse variability and repeat content of cse, pcsB, and orf1. (A) Schematic representation of the cse locus from S. thermophilus strains CNRZ368 and LMG18311. Open arrows represent ORFs and indicate their reading direction, broken arrows indicate putative promoters, and hairpin loops symbolize putative rho-independent terminators. The black arrowhead indicates the insertion site of pGh9:ISS1 within the genome of the 16D10 mutant. The percentage of sequence identity (id.) is indicated between 5′ ends, central parts, and 3′ ends of cse from CNRZ368 and LMG18311. Hatched boxes represent repeat-containing regions (var-cse). (B) Nucleic acid and amino acid sequences of var-cseCNRZ368 and the proximal region. The regions flanking var-cse are italicized. Each repeat unit is boxed, and the name of the repeat unit is indicated above the box. (C) Table of consensus repeat sequences represented in cse, pcsB, and orf1. (D) Schematic representation of repeat-containing regions. Each repeat unit is represented by a letter-containing box. Repeat units, in accordance with consensus sequences presented in panel C, are represented by capital letters, while those slightly divergent from the consensus (17% of maximum divergence) are represented by lowercase letters. Empty boxes represent regions without any repeats. Repeats in grey are common between cse from S. thermophilus strains CNRZ368 and LMG18311, orf1, and pcsB from S. thermophilus strain LMG18311. Hatched boxes represent repeats only found in cse.