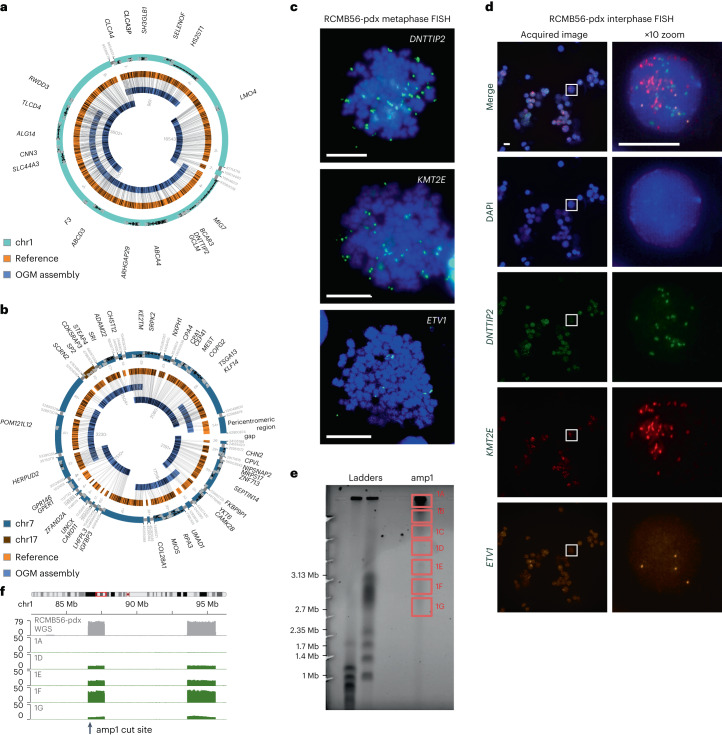
Fig. 2. Distinct high-copy extrachromosomal amplifications coexist in a SHH medulloblastoma tumor.
a, Assembly of the high-copy focal amplification amp1 from WGS and OGM of DNA from the RCMB56-pdx tumor. All breakpoints in the assembly are supported by both data types. b, Assembly of the high-copy focal amplification amp2 from the same data. All junctions are supported except a peritelomeric region adjacent to the gap, which was inferred from WGS discordant reads only. c, Metaphase FISH microscopy targeting amplified DNTTIP2 (amp1), KMT2E (amp2) and ETV1 (gain1). Images are representative of 12, 18 and 5 stained metaphase cells, respectively. Spots outside the chromosome boundaries (blue) indicate extrachromosomal amplifications. d, Interphase FISH microscopy for the same markers confirms co-amplification of all three genes on distinct amplicons. Representative image from a series of 27 images. Boxes (left) are magnified (right). Scale bars in c–d, 10 μm. e, Pulsed-field electrophoresis gel of DNA from RCMB56-pdx, generated by CRISPR-CATCH. DNA was cut using sgRNA targeting amp1, then fractionated by size through the gel. f, Sequencing coverage of the fractions indicated in e at the amp1 locus. Gray, bulk WGS; green, sgRNA targeting amp1. Amp1 is most enriched in band 1F, consistent with its 3.2 Mbp assembly length.