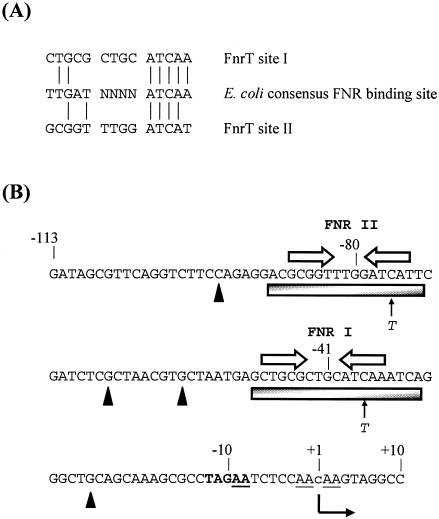
FIG. 2.
hyn regulatory region and FNR binding sites. (A) Sequences of T. roseopersicina FNR site I and FNR site II identified in this work aligned with the known consensus E. coli FNR binding site sequence. Vertical lines indicate residue identity. (B) Sequence of the FNR-regulated hyn promoter region from position −113 to position 10 relative to the putative transcription start site (position 1). The putative transcription start site is located 492 bp upstream of the hynS start codon. The initiation site, detected in an in vitro reaction, is indicated by a lowercase letter, and the direction of transcription is indicated by a bent arrow. Open arrows indicate the locations of DNA sites for FNR binding (FNR I and FNR II), while the shaded boxes show the extent of protection afforded by FNR in DNase I footprint experiments. The solid triangles indicate hypersensitive sites. The cleavage sites produced by potassium permanganate footprint analysis are underlined. The positions of single-base-pair substitutions are indicated. The −10 promoter element is indicated by boldface type. Selected positions are indicated.