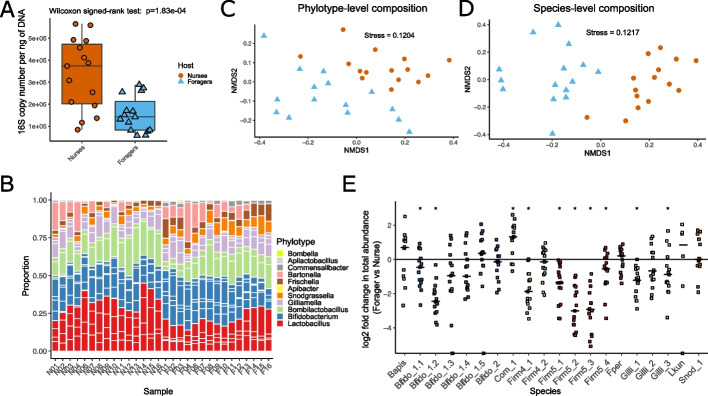
Fig. 1.
Community composition analysis of the gut microbiota of nurse and forager bees. A Boxplot of the bacterial loads of each gut represented by the number of bacterial 16S rRNA gene copies relative to 1 ng of total DNA. B Stacked bar plot of the bacterial phylotype composition for each sample. White separators within a phylotype delineate the different species. C NMDS plot of the samples’ bacterial phylotype composition. D NMDS plot of the samples’ bacterial species composition. E Log2 fold change in total normalized absolute abundance of bacterial species in forager samples relative to nurse samples from the same hive. Asterisks mark species with significant differences between nurses and foragers (Wilcoxon signed-rank test of the normalized absolute abundance of each species in the foragers against the nurses, q-value < 0.05). Data points for species of the same phylotype are shown in the same color. Horizontal bars represent medians