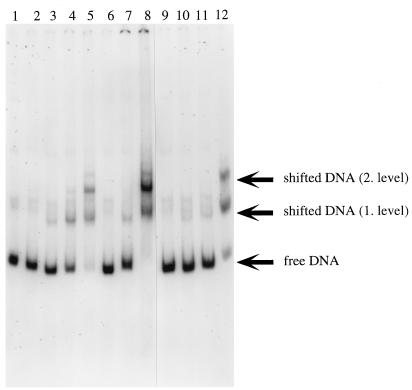
FIG. 7.
Binding of PcaU to a DNA fragment containing a portion of the pcaU-pcaI intergenic region. The probe for this gel shift experiment was the 216-bp XmnI-EcoRI fragment containing a portion of pcaU (Fig. 4). Lane 1 depicts migration of the probe without added extract. Incubation of the probe with 15 μg of protein from E. coli DH5α(pWH661+) (lane 2) or E. coli DH5α(pZR26) (lane 6) did not shift the migration pattern, whereas incubation of the probe with the same amount of protein from E. coli DH5α(pZR22) gave rise to two bands (lanes 5 and 12). Lesser amounts of protein from E. coli DH5α(pZR22) produced little or no shift: the probe had been incubated with 5.2 μg of protein for the preparation run in lanes 4 and 11; 1.5 μg of protein was provided for lanes 3 and 10; 0.15 μg of protein was provided for lane 9. The tests whose results are shown in lanes 9, 10, 11, and 12 included 1.5 mM protocatechuate in the assay. Binding of the probe to 15 μg of protein contained in an extract from E. coli DH5α(pZR22) appears to have been competitively removed by 1.8 μg of pZR23 DNA (which contains the 216-bp XmnI-EcoRI segment [lane 7]) and not by 1.8 μg of pWH661+ DNA (which is the pZR22 vector lacking the insert [lane 8]).