Fig. 5. Beta cell heterogeneity of NNAT expression is associated with changes in CpG methylation at the Nnat promoter.
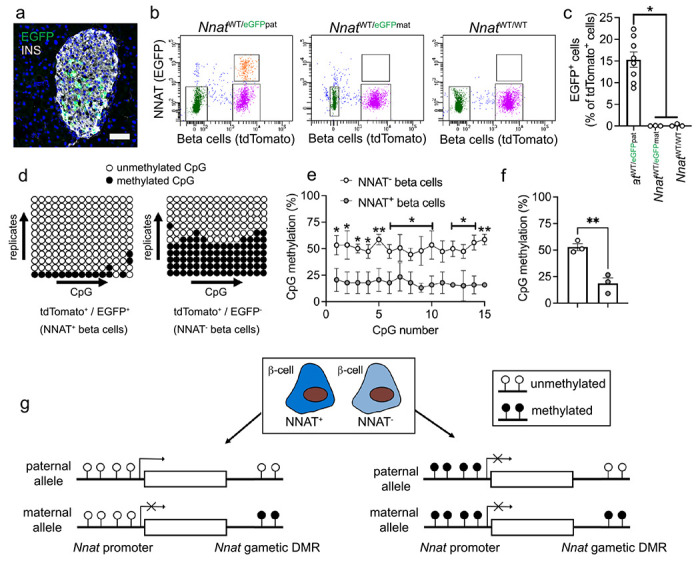
(a) Representative confocal microscopy of pancreatic cryosections from P56 mice with Nnat-driven EGFP expression from the paternal allele (NnatWT/eGFPpat) (n = 7 mice). Sections were immunostained with antibodies against EGFP (green) and insulin (INS, grey). Nuclei are visualised with DAPI. Scale bar = 50μm. (b) Separation of dispersed primary islet cells by FACS from reporter mice with insulin-driven expression of tdTomato (to label beta cells) and Nnat-driven EGFP expression from the paternal (NnatWT/eGFPpat), or maternal (NnatWT/eGFPmat) allele or wild type (NnatWT/WT) at this locus (representative image of one dispersed islet preparation, each from a single mouse) (n = 8, 3 and 3 mice, respectively, Kruskal-Wallace test with Dunn’s multiple comparisons). (c) Quantification of data in b expressed as percentage of EGFP/tdTomato co-positive primary islet cells. (d) Representative bisulphite analysis of CpG methylation at the Nnat promoter in FACS-purified islet cell populations from b (n = 3 NnatWT/eGFPpat mice with paternally expressed Nnat-driven EGFP, n = >12 clones each). Closed circles = methylated CpG, open circles = unmethylated CpG. (e, f) Quantification of data in d expressed as percentage CpG methylation across the Nnat promoter at individual CpGs (e) and across the entire Nnat promoter (f) (paired Student’s t test). (g) Schematic summarising level of CpG methylation at the Nnat promoter and gametic DMR in NNAT+ vs NNAT− beta cells. Representative images from three independent experiments and breeding pairs. * P < 0.05, ** P < 0.01.
