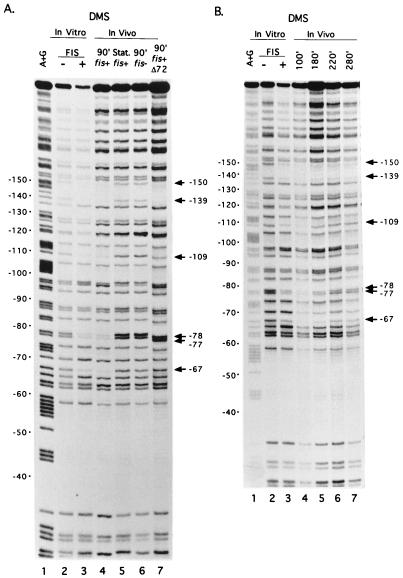
FIG. 2.
Binding of FIS to rrnBp1 in vitro and in vivo by DMS footprinting assays. (A) G residues protected by FIS in vitro and in vivo. Lane 1, A+G sequence marker; lanes 2 and 3, promoter fragment modified by DMS in vitro in the absence or presence of purified FIS (8); lane 4, wild-type promoter modified by DMS in vivo in a strain containing the wild-type fis gene (RLG911), 90 min (90′) after dilution of cells in fresh LB (exponential growth); lane 5, same as lane 4 except that cells were treated with DMS in stationary (Stat.) phase; lane 6, same as lane 4 except that cells (RLG921) contained the fis::kan767 allele; lane 7, same as lane 4 except that the rrnBp1 on the plasmid (pAJ4) contained the FIS site I mutation (Δ72; RLG912). The positions of G residues protected against DMS methylation by FIS in rrnB promoter sites I (−67, −77, and −78), II (−109), or III (−139, −150) are indicated. The degree of protection of particular positions is most easily evaluated by comparison to a reference band within the same lane (e.g., position −58, which is not protected by either FIS [8] or RNAP [38]). (B) Protection of FIS site residues in DMS footprints at different time points in a growth cycle. Plasmid DNA containing the wild-type rrnBp1 was modified in a wild-type (RLG911) strain at the indicated time point (100, 180, 220, or 280 min after dilution of a stationary-phase culture into fresh LB). Lanes 1 to 3, same as lanes 1 to 3 in panel A; lanes 4 to 7, in vivo footprints at the indicated times.