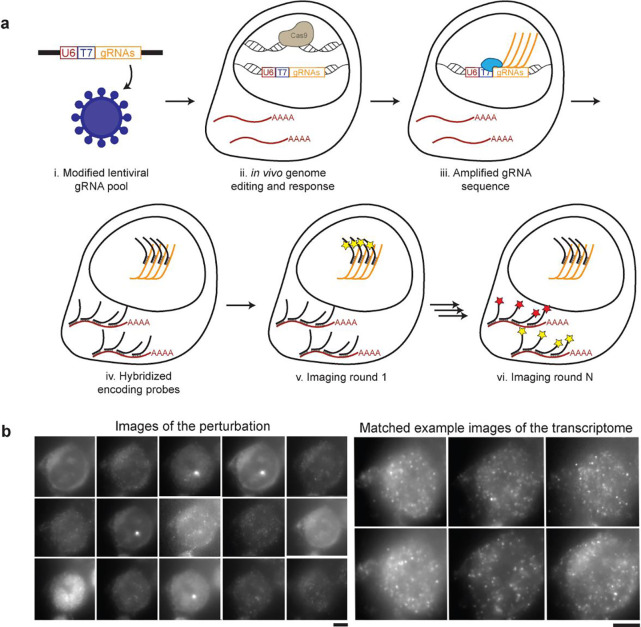
Figure 1. Perturb-FISH allows recording both gRNAs and transcriptome in cells in their spatial context.
A) Perturb-FISH workflow. i) Guides are packaged into lentiviral particles using a modified version of Lentiguide-puro that contains a T7 promoter between the end of the U6 promoter and the beginning of the guide. ii) The lentivirus is used to insert the guide sequence in the genome of the cell, resulting in genome editing. iii) T7 polymerase locally generates many copies of the gRNA in fixed cells. iv) DNA encoding probes anneal on both the target mRNA and the amplified gRNA. v) Fluorescent readouts anneal on the encoding probes, and are imaged. The fluorophores are cleaved, and this step is repeated. vi) After sequentially imaging the gRNAs, rounds of hybridization/imaging/cleavage continue to image the transcriptome in the same cells. B) Representative images of Perturb-FISH: Amplified gRNA generate a bright spot in the nuclei of cells, and the identity of gRNAs is encoded in the sequence of images in which they fluoresce (left). The transcriptome is read out the same way with MERFISH (right). T7 transcription yields higher signal amplification than the tiling of an mRNA with 30 probes, as visible by the larger size of the spots they generate. Scale bar = 10μm.