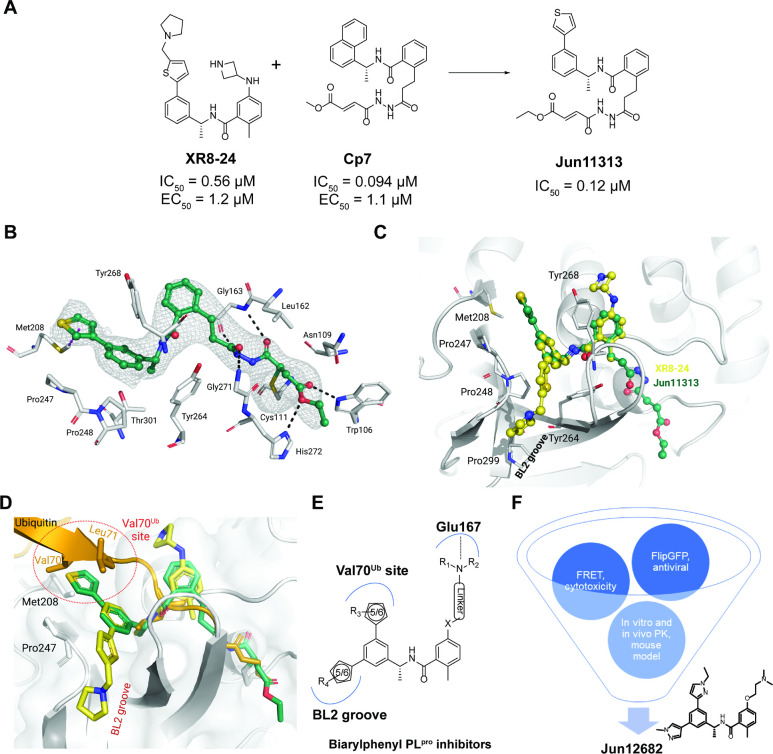
Fig. 1. X-ray crystal structure of the covalent inhibitor Jun11313 with SARS-CoV-2 PLpro and structure-based design of biarylphenyl SARS-CoV-2 PLpro inhibitors.
(A) Design of the hybrid covalent inhibitor Jun11313 based on XR8–24 and Cp7. (B) Atomic model of the Jun11313 (in green sticks and spheres) binding site in PLpro (light grey sticks, residues within a 5 Å distance of the inhibitor), with hydrogen bonds displayed as black dashed lines. Jun11313 polder map (an unbiased difference map (36)) is displayed as a grey mesh with 4σ contour. (C) Superposition of the PLpro-Jun11313 structure to the structure of the PLpro-XR8–24 complex (PDB 7LBS), with XR8–24 in yellow sticks and spheres, with the relevant residues for binding of both compounds indicated. (D) Superimposed X-ray crystal structures of SARS-CoV-2 PLpro with Jun11313 (green) (PDB: 8UVM), XR8–24 (yellow) (PDB: 7LBS), and ubiquitin (orange) (PDB: 6XAA). (E) Generic chemical structure of the designed biarylphenyl PLpro inhibitor. Critical interactions are highlighted. (F) Flow chart for the lead optimziation of PLpro inhibitors. Jun12682 was selected as the in vivo lead candidate.