Table 1.
Representative biarylbenzamide series of SARS-CoV-2 PLpro inhibitors.
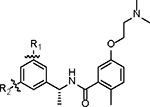
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound ID | R1 | R2 | IC50 (nM) | Ki (nM) | CC50 (μM) | FlipGFP EC50 (μM) | Antiviral EC50 (μM)* | Microsomal Stability T1/2 (min) | PDB code |
| Jun11875 |
|
|
66.2 ± 2.0 | 13.2 ± 2.0 | 4.5 ± 1.6 | --- | --- | --- | |
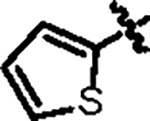
| Jun12129 |
|
|
90.9 ± 4.6 | 75.5 ± 2.0 | 8.3 ± 1.2 | --- | --- | --- | 8UUY |
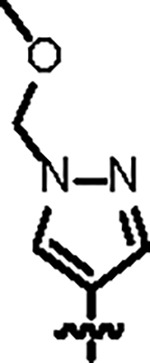
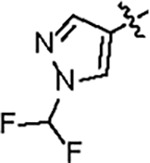
| Jun12303 |
|
|
121.5 ± 4.0 | 88.2 ± 6.0 | 56.8 ± 7.4 | 2.6 ± 0.2 | 0.32 | 136 | 8UUG |
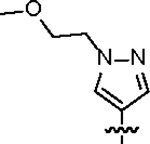
| Jun12763 |
|
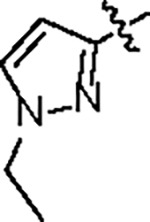
|
164.6 ± 23.0 | 37.2 ± 4.0 | > 125 | 2.0 ± 0.3 | 0.96 | --- | |
| Jun12395 |
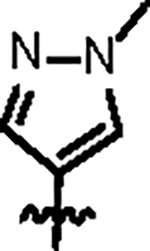
|
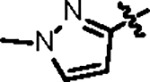
|
121.9 ± 8.0 | 47.1 ± 4.0 | 109.4 ± 16.2 | 2.9 ± 0.3 | 1.15 | 88.3 | |
| Jun12713 |
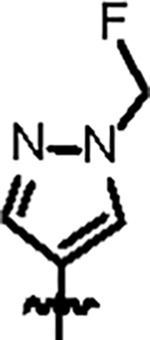
|
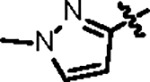
|
86.7 ± 6.0 | 34.0 ± 3.0 | 107.2 ± 6.2 | 2.7 ± 0.6 | 0.86 | --- | |
| Jun12602 |
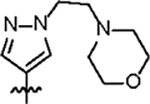
|
|
116.7 ± 9.2 | 46.9 ± 3.0 | > 125 | 14.7 ± 2.5 | 0.80 | > 145 | |
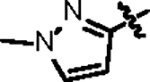
| Jun11941 |
|
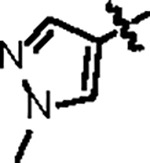
|
151.4 ± 4.0 | 34.3 ± 3.0 | 54.8 ± 8.3 | 1.8 ± 0.2 | 0.31 | > 145 | 8UUF |
| Jun12162 |
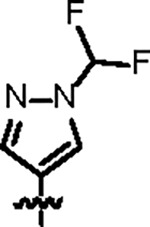
|
|
98.3 ± 7.0 | 33.6 ± 3.0 | 42.3 ± 2.3 | 2.1 ± 0.2 | 0.23 | 76.7 | 8UUU |
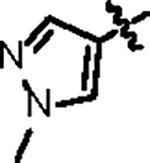
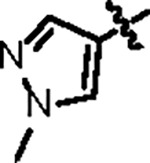
| Jun12199 |
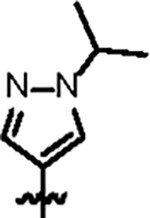
|
|
108.8 ± 10.2 | 47.6 ± 3.0 | 63.2 ± 11.3 | 0.8 ± 0.1 | 0.45 | 79.7 | 8UUH |
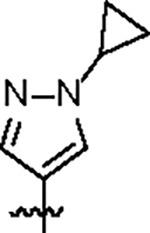
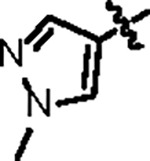
| Jun12197 |
|
|
102.7 ± 10.2 | 33.2 ± 3.0 | 43.7 ± 5.1 | 0.6 ± 0.1 | 0.35 | 116.0 | 8UUV |
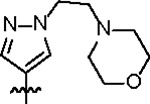
| Jun12351 |
|
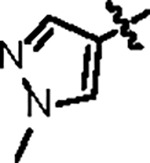
|
98.8 ± 5.2 | 29.3 ± 3.0 | > 125 | 2.0 ± 0.4 | 0.59 | > 145 | |
| Jun12603 |
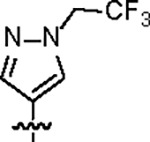
|
|
112.2 ± 6.0 | 39.8 ± 4.0 | 61.0 ± 19.3 | 2.4 ± 0.4 | 0.50 | --- | |
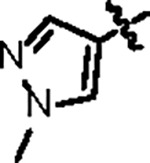
| Jun12145 |

|
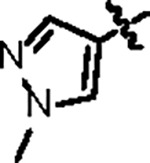
|
108.5 ± 6.2 | 35.2 ± 2.0 | 31.3 ± 5.4 | 1.3 ± 0.3 | 0.58 | 26.3 | 8UUW |
| Jun12682 |
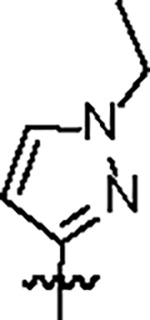
|
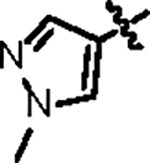
|
106.8 ± 5.0 | 37.7 ± 3.0 | 61.3 ± 4.5 | 1.1 ± 0.1 | 0.42 | 82.4 | 8UOB |
| GRL0617 |
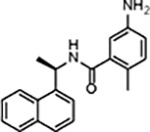
|
1918.0 ± 150.0 | 1374 ± 58 | 107.3 ± 10.9 | 22.4 ± 2.3 | > 20 | --- | 7JRN | |
The positive control nirmatrelvir had an EC50 of 0.04 μM in the SARS-CoV-2 antiviral assay.
