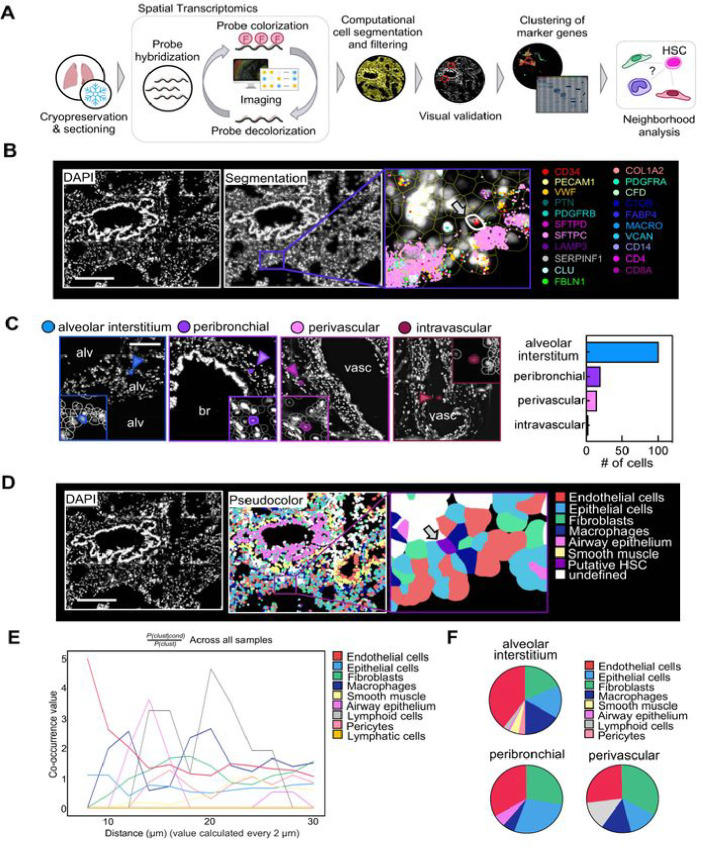
Figure 4.
Spatial mapping of phenotypic CD34+ HSPCs in the lung. (A) Spatial transcriptomics analysis workflow. smFISH was performed to visualize gene expression in human lung tissue. Transcripts were assigned to individual cells after cell segmentation and putative HSPCs were computationally identified based on their gene signature and visually validated. Lung stromal and immune cells were annotated based on marker gene clusters (Figure S8). Subsequently, co-occurence analysis was performed. (B) Representative image of a putative HSPC in its pulmonary niche. Upper panel (left to right): DAPI staining, QuPath segmentation, zoom on putative HSC (arrow). Selected transcripts are shown. Scale bar, 250 μm (C) Anatomic location of candidate cells in the lung. Representative images of phenotypic HSPCs in four major locations (alveolar interstitium, peribronchial, perivascular or intravascular) and proportion of cells in each location. Alv, alveolar space; br, bronchus; vasc, vasculature. Scale bar, 150μm. (D) Pseudo-coloring of cell types in the lung tissue based on marker clustering (Figure S8). Zoom on putative HSPC in niche. Scale bar, 250 μm. (E) Squidpy co-occurrence score computed every 2 μm between putative HSPCs and the rest of the clusters across lung tissue sections from 4 organ donors. High score values indicate greater co-occurrence probability; endothelial cells (red) and macrophages (dark blue) co-occur with the HSPCs at short distances. (F) Pie graphs showing the proportion of neighboring cells within a radius of 20 μm from the putative HSPCs in the major anatomic locations.