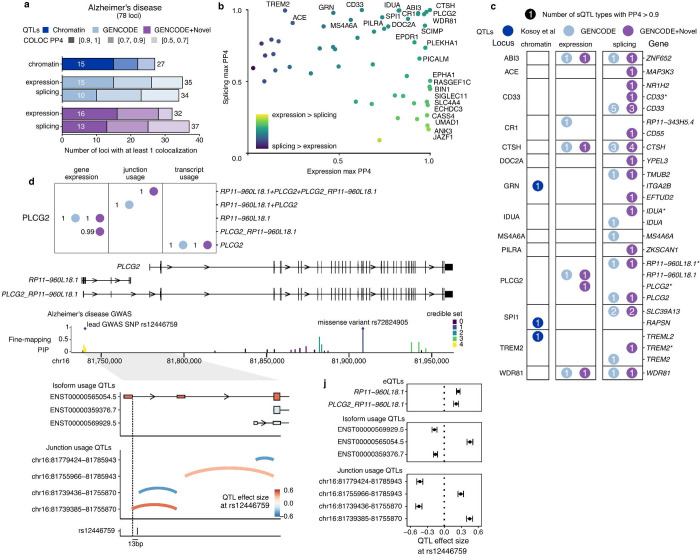
Fig. 5 |. Novel isoforms improve interpretation of Alzheimer’s disease colocalization.
a, Colocalization discovery rate for microglia chromatin, expression and splicing QTLs in Alzheimer’s disease at different thresholds of colocalization posterior probability (PP4). “Splicing” represents a combination of all 8 sQTL modalities. b, Comparing the maximum expression and splicing PP4 for each locus across both references. Labels refer to loci with PP4 > 0.9. c, Colocalization results at each locus in Alzheimer’s disease. Numbers within circles refer to the number of different sQTL modalities with a colocalization PP4 > 0.9. Asterisks refer to additional genes being implicated in fusion isoforms and/or overlapping splice junctions. d, Multiple QTLs colocalize with the PLCG2 Alzheimer’s disease risk locus. The lncRNA RP11–960L18.1 upstream of PLCG2 produces fusion isoforms detectable as novel isoforms in microglia. Fine-mapping of the PLCG2 locus identifies a credible set of SNPs nearby the first exon of RP11–960L18.1, distinct from the rare missense mutation in the gene body of PLCG2. Transcript usage QTLs colocalizing with the GWAS variant show increased usage of the upstream TSS. i, Junction usage QTLs identify changes in splicing around the upstream fusion TSS and decreased usage of the downstream PLCG2 TSS. j, Quantified effect sizes of each QTL type relative to the lead GWAS SNP rs12446759.