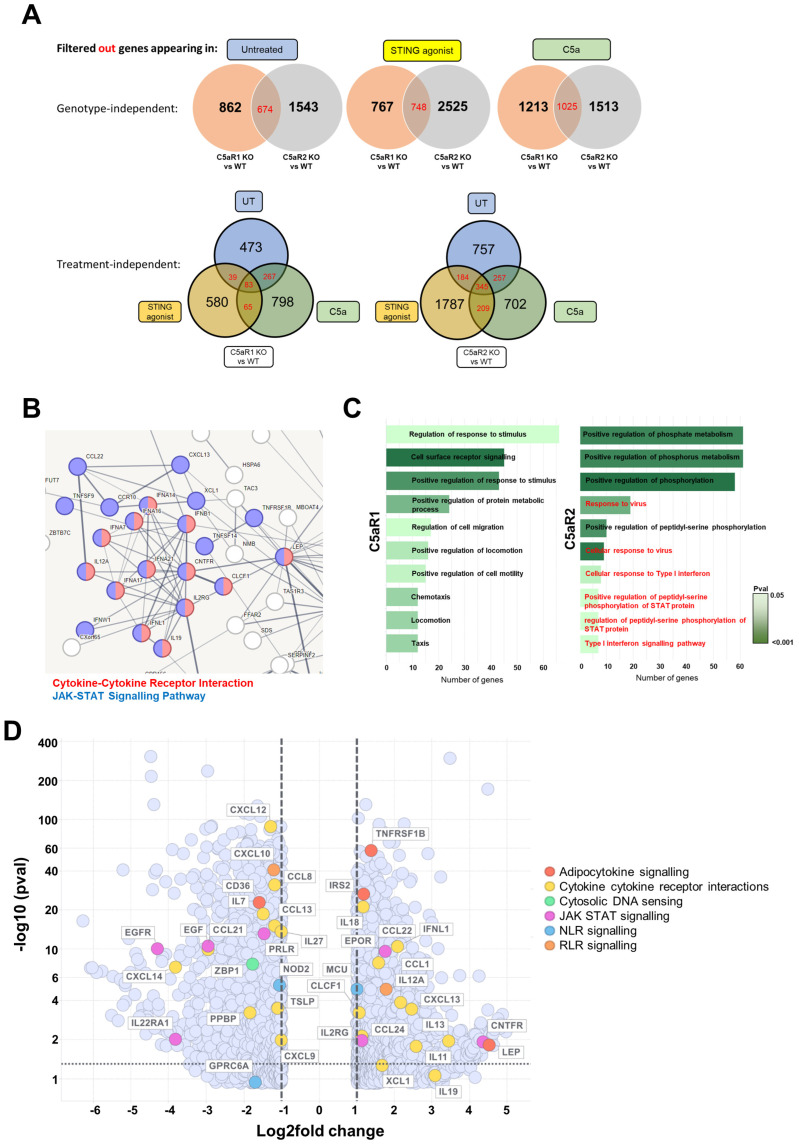
Figure 5.
Pathway analysis of transcriptomic data reveals an enhanced antiviral phenotype in C5aR2 KO THP-1 cell lines compared to WT. (A) Unique, genotype-dependent, treatment-dependent significantly regulated genes (p ≤ 0.05, log2FC < ±1) were identified. (B) An unbiased cluster analysis and pathway analysis was performed using STRING, and the significantly enriched pathways were highlighted in red (Cytokine-Cytokine Receptor Interaction) or blue (JAK-STAT Signalling Pathway) (Supplementary Figures S6–S9). (C) An unbiased pathway analysis was performed using PANTHER and GO Biological Process gene sets. Red text highlights relevant anti-viral pathways (D) Significantly regulated KEGG gene sets from the GSEA were manually triaged, selecting for unique pathways for each condition, genotype-specific stimulation-independent pathways, or genotype-independent stimulation-independent pathways, filtering out stimulation-specific genotype-independent pathways, or pathways common to all conditions. Significantly regulated inflammatory pathways from the C5aR2 KO vs. wild-type comparison (C5a or STING agonist-treated) were manually curated. Selected genes from KEGG gene sets were marked in a volcano plot for STING agonist-treated C5aR2 KO cells vs. STING agonist-treated wild-type cells.