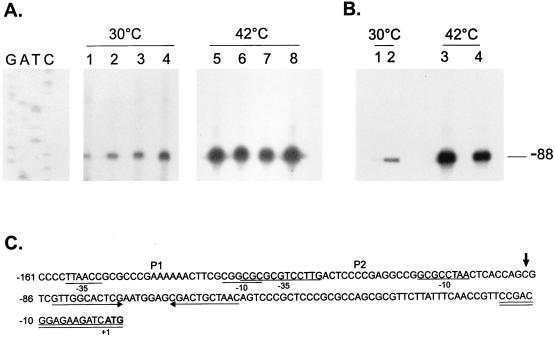
FIG. 10.
Primer extension mapping of the transcription start sites in hrcA and CIRCE mutants. (A) An 18-nucleotide primer complementary to nucleotides −15 to +3 was 5′ end labelled and hybridized to 50 μg of total RNA isolated from NA1000 cells harboring transcription fusion pMA11 (lanes 1 and 5), pRB19 (lanes 2 and 6), pRB20 (lanes 3 and 7), or pRB21 (lanes 4 and 8) grown at 30°C or exposed to 42°C for 30 min. The hybrids were then extended by using reverse transcriptase, and the extension products were resolved by denaturing gel electrophoresis and autoradiography. (B) The same procedure as described above for panel A was performed except that the RNA used was isolated from NA1000 (lanes 1 and 3) or LS2293 cells (lanes 2 and 4) grown at 30°C or after a 15-min exposure to 42°C. The sequencing ladder shown in panel A was generated by using the same 18-residue oligonucleotide as the primer and the plasmid pUC19 containing the groESL regulatory region as the template. (C) Sequence of the 5′ region of the groESL operon. The transcription start site is shown by an arrow over the nucleotide sequence. The putative −10 and −35 regions of the P1 and P2 promoters are underlined. The CIRCE element is depicted by arrows under the nucleotide sequence. The oligonucleotide used in primer extension experiments is complementary to the sequence underlined twice. The ATG of the initiator methionine is shown in bold type.