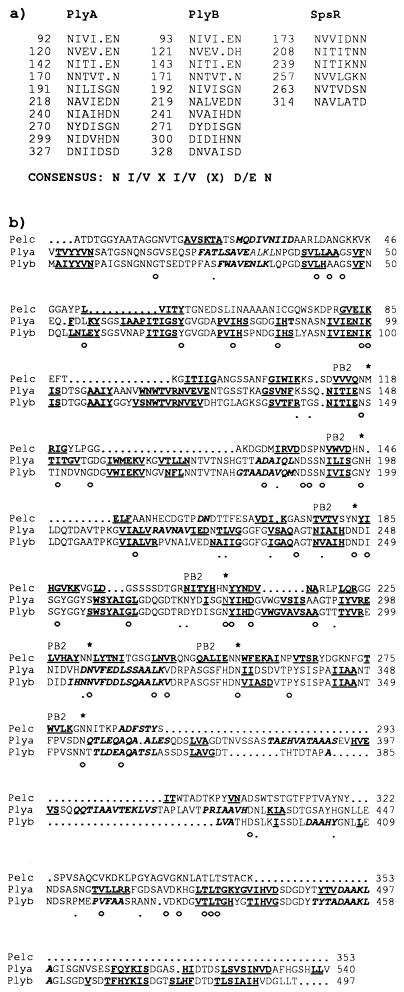
FIG. 5.
(a) Repeat motifs found in PlyA, PlyB, and SpsR. The position in the sequence of the first amino acid in each repeat is indicated. A rough consensus sequence is shown. PlyA and PlyB each contain 10 copies of the motif; SpsR contains 6 copies. (b) Sequence alignment of PelC from E. chrysanthemi with PlyA and PlyB from R. leguminosarum bv. viciae, showing conservation of secondary structure. The mature form of PelC (lacking the N-terminal signal peptide) was used for the alignment. Residues that are conserved in all three proteins (○) and between PelC and either PlyA or PlyB (·) are indicated beneath the alignment. Structural elements are indicated in boldface type, with α-helices in italics and β-strands underlined. The secondary structure of PelC is known from the crystal structure (49). The β-strands that form the PB2 β-sheet in PelC are indicated. The residues which form stacks stabilizing the parallel β-helix of PelC are indicated by asterisks above the sequence. The stacks are composed mainly of Asn residues, which occur as the second residue after the end of the PB2 β-strands. The secondary structure of PlyA and PlyB was predicted by PHDsec (34) and aligns well with the known structure of PelC, suggesting that these proteins may have a similar fold.