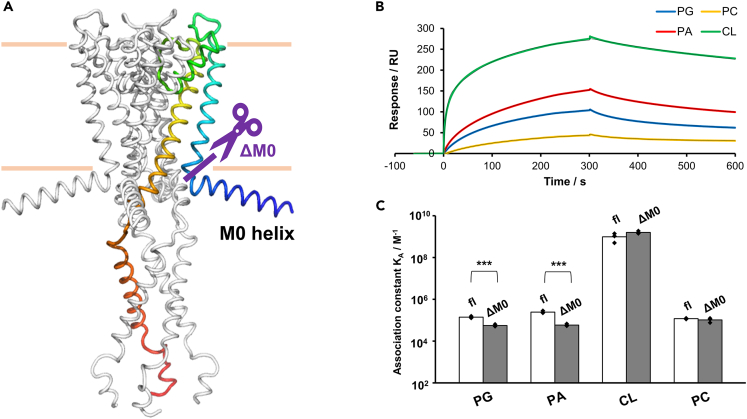
Figure 4.
Effect of truncation of the M0 helix on the affinity of lipids toward KcsA
(A): Location of deletion on KcsA. ΔM0-KcsA is the mutant in which the N-terminal M0-helix domain was truncated (purple line).
(B): SPR sensorgrams showing the interactions of PG, PA, CL, and PC with ΔM0-KcsA immobilized on the C6-SAM modified sensor chip; 100 μM of the lipids dissolved in acidic buffer (pH 4.0) were injected at a flow rate of 30 μL min−1. Colored lines indicate experimentally obtained sensorgrams, while black lines indicate fitted curves.
(C) Affinity of PG, PA, CL, and PC toward fl-KcsA and ΔM0-KcsA in acidic buffer (pH 4.0). Each affinity was calculated from the sensorgrams by fitting to a 1:2 heterogeneous ligand binding model, as listed in Table S4. Asterisks denote statistical significance (∗∗∗p < 0.001). Data presented as mean ± SD (n = 3) are given in Table S4.