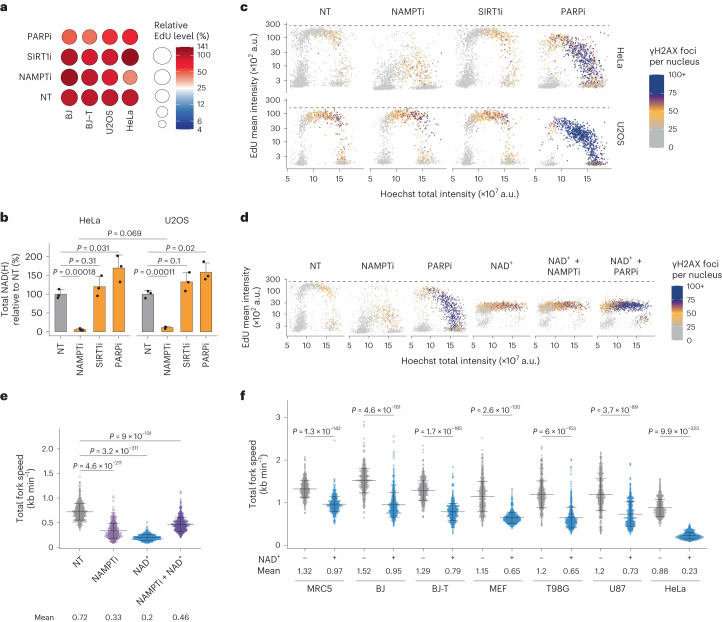
Fig. 1. Exogenous NAD+ impairs DNA replication.
a, Flow cytometry analysis of EdU incorporation in S-phase cells relative to the non-treated (NT) condition. Cells were treated for 24 h with 10 nM NAMPTi, 2 µM SIRT1i or 10 µM PARPi. The experiment was performed twice with similar results. b, NAD(H) quantification in HeLa and U2OS cells treated as in a. Data are mean + s.d., n = 3 biological replicates, two-tailed Student’s t-test. c, QIBC analysis of γH2AX foci, EdU intensity and DNA content in HeLa and U2OS cells treated as in a. Representative experiment from three independent experiments. d, QIBC analysis of γH2AX foci, EdU intensity and DNA content in HeLa cells treated for 24 h without (NT) or with 10 nM NAMPTi, 10 µM PARPi and/or 2 mM NAD+. Representative experiment from three (NT, PARPi and NAD+) and two (all other conditions) independent experiments. e, Replication fork speed in HeLa cells treated for 24 h without (NT) or with 10 nM NAMPTi and/or 2 mM NAD+. Data are mean ± s.d., means are indicated. Scored forks: NT, 553; NAMPTi, 515; NAD+, 552; NAMPTi+NAD+, 557. Two-tailed Welch’s t-test. f, Replication fork speed in different cell lines treated for 24 h without or with 2 mM NAD+. Data are mean ± s.d., and mean values in kb min−1 of the fork speed are indicated at the bottom of the plot. Scored forks: MRC5 NT, 525; MRC5 NAD+, 535; BJ NT, 543; BJ NAD+, 523; BJ-T NT, 530; BJ-T NAD+, 542; MEF NT, 529; MEF NAD+, 612; T98G NT, 519; T98G NAD+, 519; U87 NT, 522; U87 NAD+, 539; HeLa NT, 543; HeLa NAD+, 582. Two-tailed Welch’s t-test. Replication forks were scored for two biological replicates. Numerical data are available in source data files.