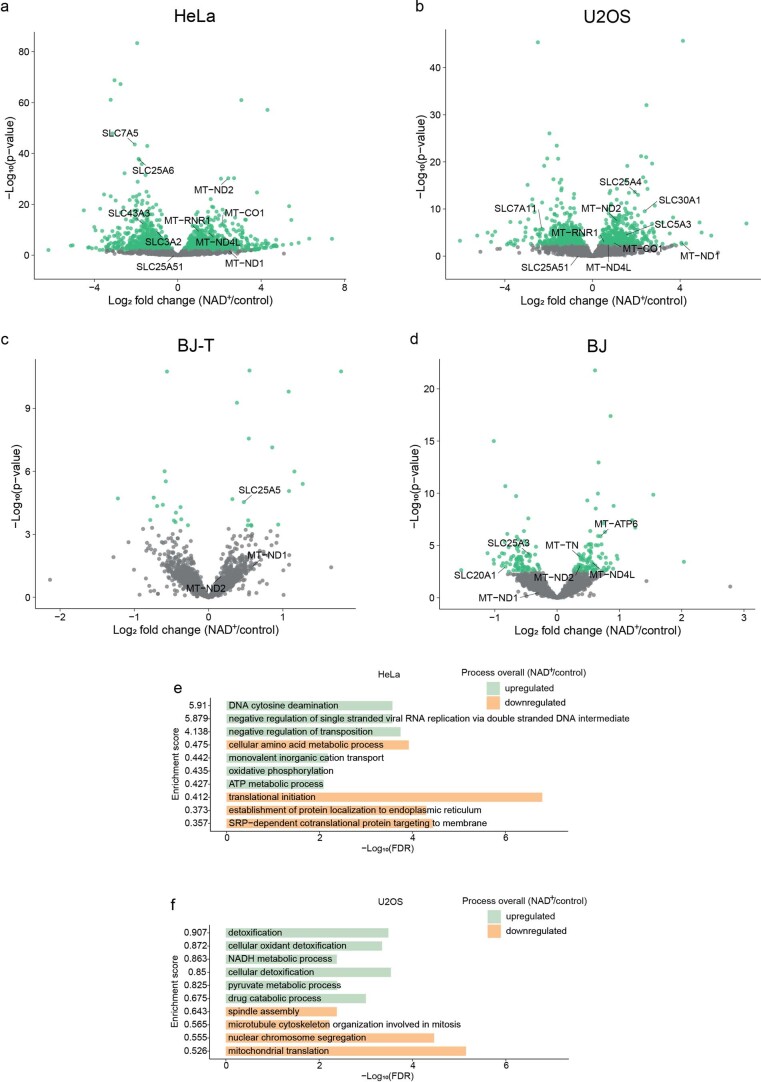
Extended Data Fig. 4. RNA-seq analysis of NAD+-treated cells.
a-d, Volcano plots of differentially expressed genes after treatment for 24 h with 2 mM NAD+ in HeLa (a), U2OS (b), BJ-T (c), and BJ (d) cells. n = 3 independent biological samples per condition sequenced once with multiplexing. Green color indicates significant changes (FDR-adjusted p-value < 0.05), non-significant changes are in grey. SLC-, solute carrier genes; MT-, mitochondrial genes. e-f, Top 10 most enriched biological processes (FDR < 0.01) from STRING (Functional Enrichment Analysis) affected by 2 mM NAD+ for 24 h treatment from RNA-seq analysis of HeLa (e) and U2OS (f) cells. Numerical data are available in Source Data Files.