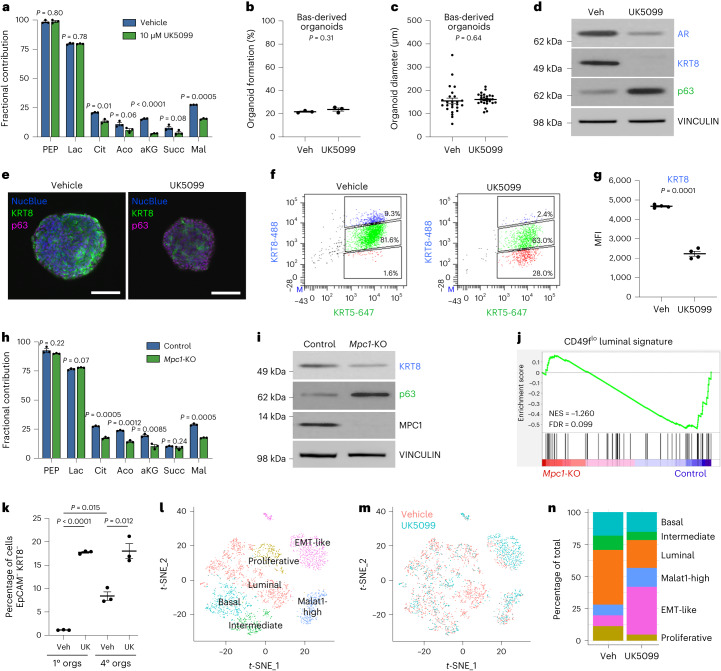
Fig. 3. Inhibition or knockout of the MPC prevents basal-to-luminal differentiation.
a, [U-13C]glucose tracer analysis of vehicle- and 10 μM UK5099-treated basal-derived organoids 7 d after plating (n = 3 independent biological replicates). Data are shown as mean ± s.e.m. b,c, Percentage organoid formation (n = 3 independent biological replicates) (b) and organoid diameter (n = 25 independent biological samples) (c) of vehicle- and 10 μM UK5099-treated basal-derived organoids 7 d after plating. d, Western blot analysis of luminal markers androgen receptor (AR) and KRT8 and basal marker p63 in vehicle- and 10 μM UK5099-treated basal-derived organoids 7 d after plating. e, Immunofluorescence of luminal marker KRT8 and basal marker p63 in representative vehicle- and 10 μM UK5099-treated basal-derived organoids 7 d after plating. Scale bars, 100 μm. f, Intracellular flow cytometry of KRT8 and basal marker cytokeratin 5 (KRT5) in vehicle- and 10 μM UK5099-treated basal-derived organoids 7 d after plating. g, Quantification of mean fluorescence intensity (MFI) of KRT8 from panel f (n = 4 independent biological replicates). h, [U-13C]glucose tracer analysis of control and Mpc1-KO basal-derived organoids (n = 3 independent biological replicates). Data are shown as mean ± s.e.m. i, Western blot analysis of basal and luminal markers in control and Mpc1-KO basal-derived organoids. j, GSEA showing negative enrichment of CD49flow luminal signature18 in Mpc1-KO relative to control basal-derived organoids. k, Flow cytometry quantification of percentage of EpCAM−KRT8− cells in vehicle- and 10 μM UK5099-treated primary and quaternary basal-derived organoids (n = 3 independent biological replicates). l, t-Distributed stochastic neighbour embedding (t-SNE) plot of scRNA-seq data on quaternary prostate organoids illustrating distinct cell populations. m, t-SNE plot of vehicle- and 10 μM UK5099-treated cells from scRNA-seq data. n, Quantification of percentage of vehicle- and 10 μM UK5099-treated cells in each cluster from scRNA-seq data. For all panels, error bars represent s.e.m. P values were calculated using an unpaired two-tailed t-test with Welch’s correction. Aco, aconitate; Cit, citrate; EMT, epithelial–mesenchymal transition; Lac, lactate; Mal, malate; orgs, organoids; Succ, succinate; UK, UK5099; Veh, vehicle.