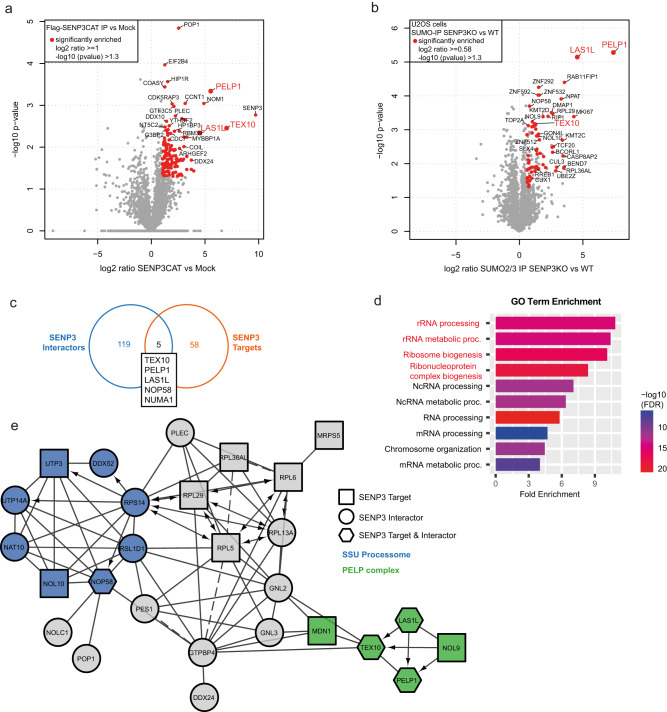
Fig. 1. The network of SENP3 interactors and targets.
a Volcano plot of quantitative MS analysis after anti-FLAG-IP of HEK923T cell lysates transiently expressing FLAG-SENP3C532S. Significantly enriched interactors are color-coded (log2 ratio ≥ 1, –log10 p value ≥ 1.3). Identification of candidates is based on two-sided Student’s t-test analysis comparing LFQ intensities of IPs from FLAG-SENP3 expressing and control cells. Members of the PELP1 complex are additionally highlighted. Experiments were performed in triplicates. b Volcano plot summarizing the results of a quantitative MS analysis comparing SUMO targets of U2OS WT and U2OS SENP3 KO cells. Proteins were enriched by anti-SUMO2/3 IP. Hits considered as significant are highlighted in red (log2 ratio ≥ 0.58, –log10 p value ≥ 1.3). Candidate SENP3 target proteins were defined by two-sided Student’s t-test analysis comparing LFQ intensities of anti-SUMO2/3 IP with the respective IgG control IPs. Components of the PELP1 complex are additionally highlighted. Experiments were performed as triplicates. c Venn diagram representing the overlap of proteins identified as SENP3 interactors (a) or SENP3 targets (b). d Gene Ontology term enrichment analysis of biological processes (GO BP) of high-confidence SENP3 interactors (a) and targets (b) identified by MS (interactors: log2 ratio ≥ 1, targets: log2 ratio ≥ 0.58). Shown here are the top 10 enriched biological processes. Highlighted in red are terms directly connected to the process of ribosome biogenesis. The enrichment analysis was done using the ShinyGO tool, applying an FDR cutoff of 0.05. e The interconnection of significantly enriched SENP3 interactors and targets (a, b) visualized as STRING network. The minimum required interaction score was set to high confidence (0.7). Proteins that are part of the PELP1 complex are color-coded in green, those being part of the SSU processome are marked in blue. Only connected proteins are visualized.