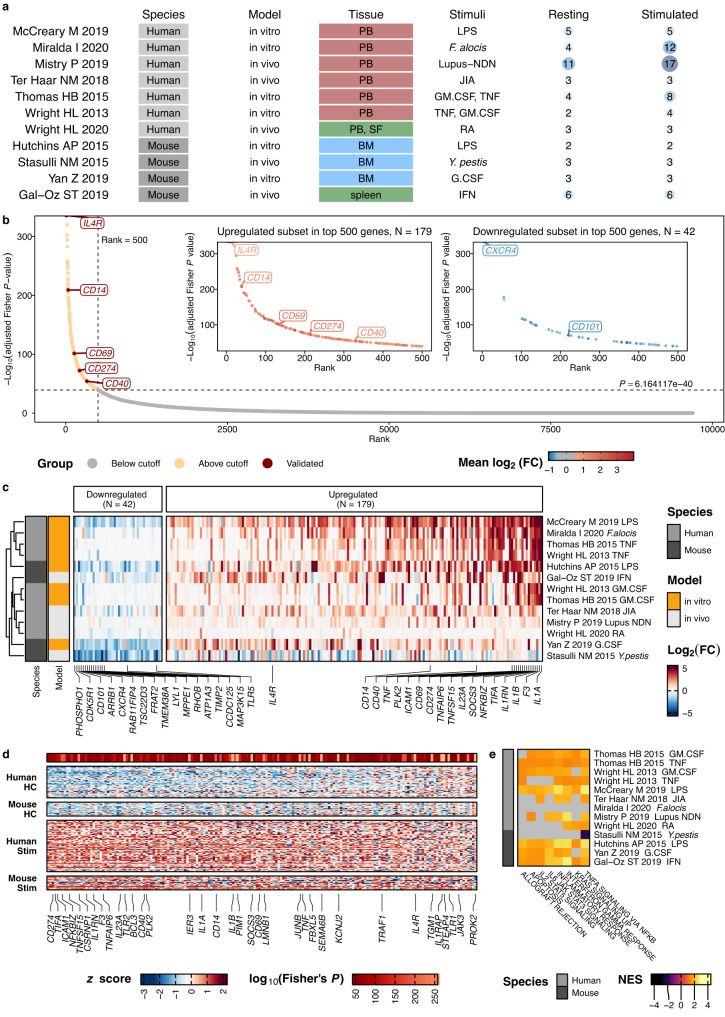
Fig. 3. A core inflammation program is conserved across mouse and human neutrophils.
a Overview of the 11 studies integrated for analysis. Differential expression testing was performed independently for each study, resting neutrophils within each study were used as control. b A combined analysis of the neutrophil response to activation/inflammation identifies 179 consistently upregulated (core inflammation program) and 42 downregulated genes in inflammation. Shown are all (N = 9697) tested genes ranked by their −log10 Benjamini-Hochberg adjusted Fisher P-value (adjusted Fisher’s combined test on two-sided P-values from individual differential expression analyses for each comparison). The 500 genes with the lowest P-values were subjected to an additional filtering step based on a log2 fold change cutoff ≥0.5 and ≤−0.5 for upregulated and downregulated genes. Highlighted (IL4R, CD14, CD69, CD274, CD40) upregulated genes were validated experimentally (Figs. 6 and 7). c 42 genes downregulated in inflammation and 179 core inflammation genes are shared across studies. Shown are the log2 fold changes across comparisons of genes up- and downregulated in inflammation. Rows represent a comparison, columns represent genes that passed our meta-analysis thresholds. Columns are arranged by the mean log2 fold change across all comparisons. For each direction, the 15 genes with the highest absolute log2 fold change are labeled, as well as genes encoding for proteins validated in Figs. 6 and 7. d Core inflammation genes are not expressed in resting neutrophils in both species and are induced upon activation. Shown is a heatmap with relative expression values (z-score for each gene across samples) of the core inflammation genes. Each column represents a gene, and each row a sample. P-values: Results of a Benjamini-Hochberg adjusted Fisher’s combined test. We labeled the top 20 genes with the lowest P-values, genes that were also labeled in (c), and manually labeled TRAF1 and JUNB. e Conserved Gene Set Enrichment Analysis based on rankings derived from each comparison’s log2 fold changes. Heatmap showing normalized enrichment scores. Only pathways that have been significant in more than 50% of comparisons are depicted. Gray fields indicate nonsignificant NES values. Source data are provided as a Source Data file.