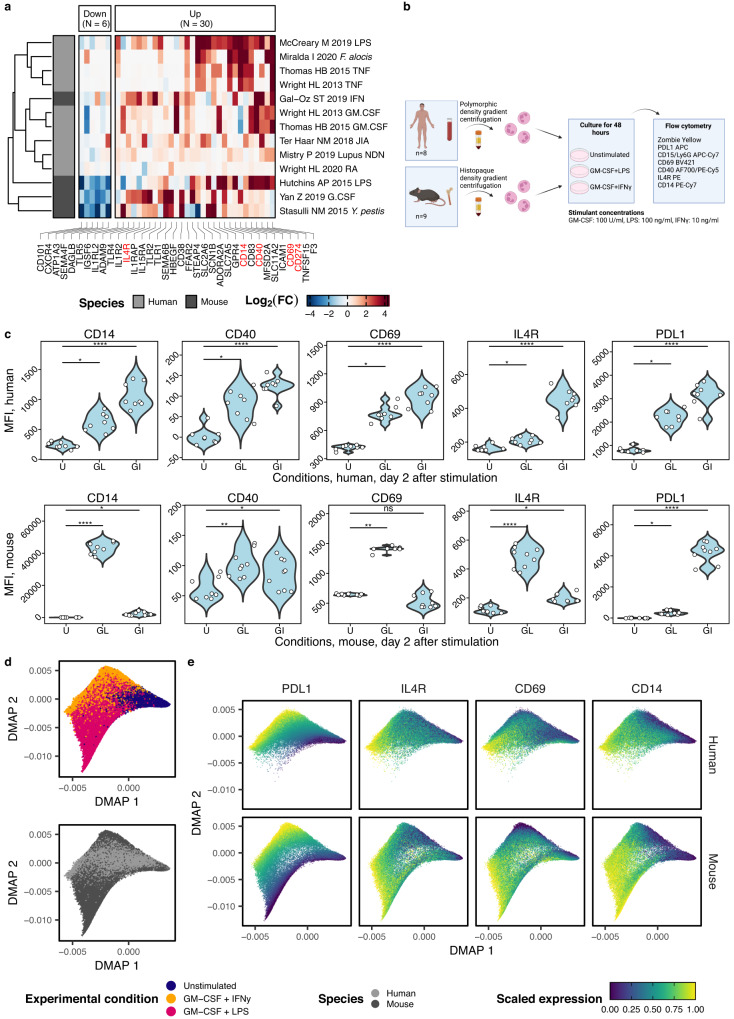
Fig. 6. Experimental validation of the core inflammation program on the protein level.
a Inflammation-specific protein-coding genes were identified by filtering the genes depicted in Fig. 3c for the surfaceome as previously described45. Genes that encode proteins selected for validation (based on antibody availability and panel design) are labeled in red. b Experimental overview. Human and mouse neutrophils were isolated from peripheral blood or bone marrow, respectively, cultured 48 h, and analyzed in flow cytometry. c Flow cytometry analysis of resting and activated mouse and human neutrophils. The gating strategy is depicted in Supplementary Fig. 11. Significance indicates adjusted P-values of a Dunn’s test (two-sided) that followed a Kruskal-Wallis H test (significant for all markers). Exact P-values are provided in the Source Data file. d Top, diffusion map embedding of neutrophils from humans and mice cultured for 48 h with or without GM-CSF + LPS and GM-CSF + IFN-γ. Diffusion map embedding calculated based on CD69, CD14, IL-4R, and PD-L1. Bottom, diffusion map embedding of human and mouse neutrophils, colored by species. e Diffusion map embedding colored by marker expression highlights a continuum driven by increasing expression of the activation markers. Source data are provided as a Source Data file.