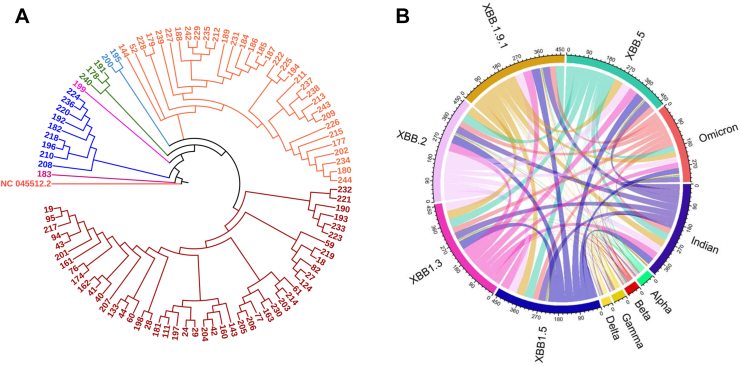
Fig. 3.
(A) The FASTA sequences used in the analysis are acquired from GenBank®. All the sequences are from the XBB variant, and we were interested in finding the variations present in the S gene. All the sequences were mapped to the reference Wuhan-Hu 1 SARS-CoV-2 isolate’s complete genome (NC_045512.2). Sequence analysis was performed using Genome Annotation Transfer Utility (GATU), an ‘in house’ bioinformatics pipeline written in Python (available upon request). This pipeline was developed to rapidly process and translate the sequences. The sequences were cropped to the gene of our interest using coordinates provided by NCBI and translated to protein sequences automatically through the ExPasy server. After translation, all the sequences were mapped with the reference protein sequence using the MAFFT (ver.7.0) multiple sequence alignment program Phylogenetic tree was also constructed using NJ method and MEGA X program with Maximum Likelihood approach. The variation was then called out and deposited as a data file for further interpretation. (B) The chord diagram shows the common mutations among different variants of SARS-CoV-2. Majority of mutations are common among XBB sequences (34–40), whereas 2–3 mutations are common between the alpha, beta, gamma and delta variants and the remaining Omicron sub-variants (the details of common mutations are presented in Table 2). Footnotes: FASTA, FastAll; NCBI, GATU, Genome Annotation Transfer Utility; MAFFT, Multiple Alignment using Fast Fourier Transform; MEGA X, Molecular Evolutionary Genetics Analysis software; NCBI, National Center for Biotechnology Information; NJ, Neighbour-Joining.