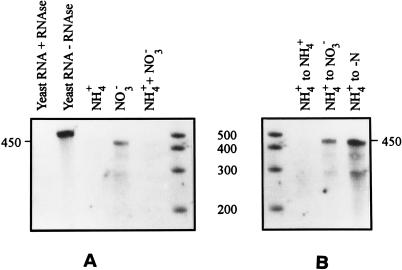
FIG. 3.
RPA showing ntcA transcript levels in Synechococcus sp. strain WH 7803 grown on different nitrogen sources. (A) The internal RNA probe was hybridized to 8 μg of total RNA extracted from cultures grown on ASWNH4 (NH4+), ASWNO3 (NO3−), and ASWNH4+NO3 (NH4+ + NO3−). Yeast RNA subjected to the RPA protocol (first lane) showed that the probe is degraded entirely when no homologous transcript is present, and yeast RNA subjected to the RPA protocol without RNase digestion (second lane) showed that the probe remained intact throughout the procedure. The last lane contains single-stranded-RNA standards. (B) The internal RNA probe was hybridized to 8 μg of total RNA extracted from cultures transferred from ASWNH4 and acclimated for 20 h in ASWNH4 (NH4+ to NH4+), ASWNO3 (NH4+ to NO3−), and ASW0 (NH4+ to −N). The first lane contains single-stranded-RNA standards. In both panels, the full length (in bases) of the transcript-probe duplex is indicated. This is shorter than the undigested probe which contains flanking regions of the cloning vector. The sizes of single-stranded-RNA standards are shown between the panels (in bases).