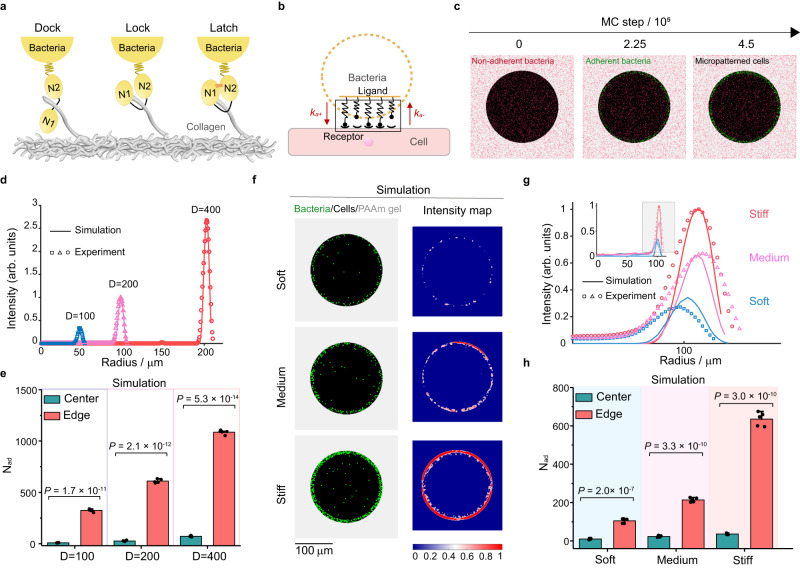
Fig. 4. Monte Carlo simulations of bacterial adhesion onto micropatterned host cell monolayers.
a Schematic diagram of DLL mechanism. N1 and N2 were two subdomains of Cna that wrapped collagen molecules (gray) through conformational changes. b Schematic diagram of bacteria–cell adhesion at the single-cell level. denoted the association rate whereas represented the dissociation rate. c Typical simulation results characterizing the bacteria–cell adhesion under geometric constrains at different Monte Carlo steps (MC step). Non-adherent bacteria were marked as red solid squares, and adherent bacteria were labeled as green solid squares. Micropatterned cells were denoted with a black background. At least three independent experiments were performed in each specific situation. d Comparison of average fluorescence intensities of adherent bacteria along the radial direction obtained from the simulations and the corresponding experiments. The diameter of the simulated host cell monolayers was set as 100, 200, and 400 μm, respectively. e Simulation data showing significant differences in the number of adherent bacteria (Nad) onto the center and edge of the circular cell monolayers with different diameters (100, 200, and 400 μm). All the simulated data were displayed as mean ± SD (n = 5; two-tailed unpaired t-test). f Simulation results presenting spatial distributions of adherent bacteria onto the host cell monolayers cultured on soft (10.14 kPa), medium (32.29 kPa) and stiff (93.46 kPa) substrates, where the adherent bacteria were labeled as green solid squares, the micropatterned cell monolayers were marked with a black background, and the underlying PAAm hydrogel substrates were shown in gray. g Comparison of average fluorescence intensities of adherent bacteria along the radial direction obtained from the simulations and the corresponding experiments, in which the diameter of the circular cell monolayers was 200 μm. h Simulation data showing significant differences in the number of adherent bacteria onto the center and edge of the circular cell monolayers cultured on soft, medium and stiff substrates (n = 5; two-tailed unpaired t-test). All representative data were repeated at least three times with similar results. Source data are provided as a Source Data file.