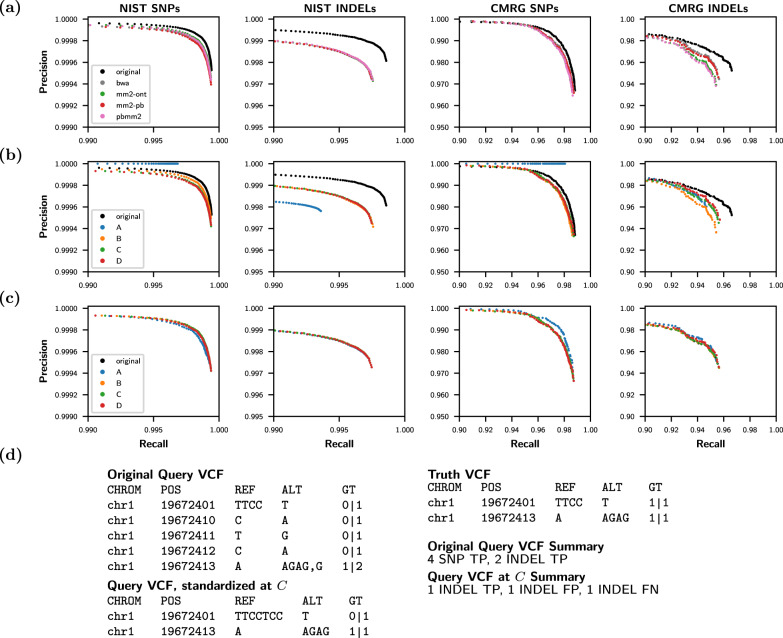
Fig. 3. vcfeval baseline precision and recall.
vcfeval precision-recall plots for Truth Challenge V2 submission K4GT3 on the NIST whole genome and Challenging Medically Relevant Genes (CMRG) datasets for single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) separately. a Evaluating the original query variant call file (VCF) and after changing the query variant representation using the alignment parameters of common aligners (see Fig. 2). b Evaluating the original query VCF and after changing the query variant representation to design points A, B, C, and D (see Fig. 2). c Standardizing the five representations from (b) using vcfdist prior to evaluating with vcfeval improves consistency. d A real example demonstrating why the original K4GT3 query VCF appears to significantly outperform other representations in (a) and (b). Each VCF shows the variant chromosomes (CHROM) and positions (POS) in addition to the reference (REF) and alternate (ALT) alleles and their genotypes (GT). Because vcfeval discards query phasing information and allows any possible local phasing, the original fractured variant representation is considered entirely correct (all true positives) whereas the more succinct standardized representation at C is not (it contains false positives and false negatives). Source data are provided as a Source Data file.