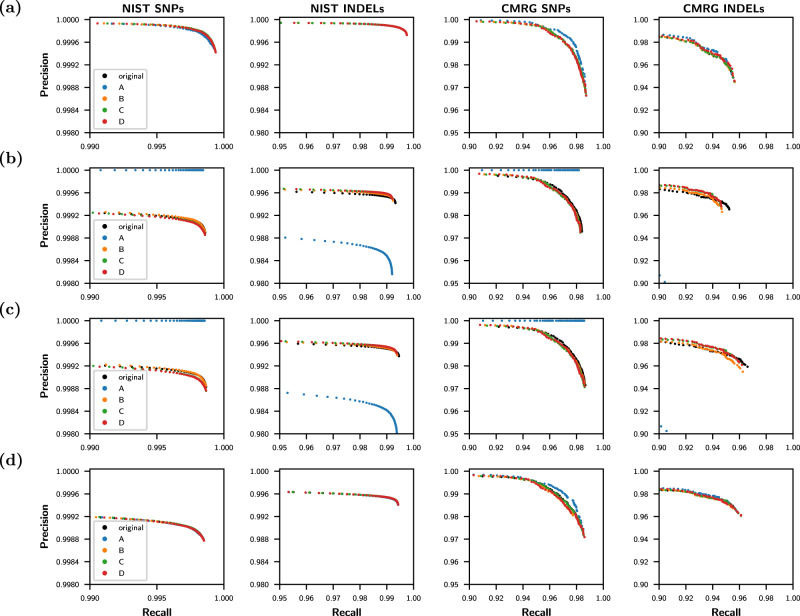
Fig. 4. vcfdist precision and recall.
a Standardized vcfeval and (b–d) vcfdist precision-recall plots for Truth Challenge V2 submission K4GT3 on the NIST whole genome and Challenging Medically Relevant Genes (CMRG) datasets, for single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) separately. For all plots, the original query VCF is evaluated before and after changing its variant representation to design points A, B, C, and D (see Fig. 2). a This plot is identical to Fig. 3c, but using axes consistent with the remainder of this figure. b Evaluation with vcfdist, turning the options for standardization and partial credit off; this can be directly compared to vcfeval’s performance in Fig. 3b. Note that the original representation no longer outperforms other representations, since local phasing is enforced. c Evaluation with vcfdist, allowing partial credit but not standardization, resulting in minor CMRG recall improvements. d Evaluation with vcfdist, allowing partial credit and standardizing variant representation. This results in the most consistent results, and is the recommended usage. Source data are provided as a Source Data file.