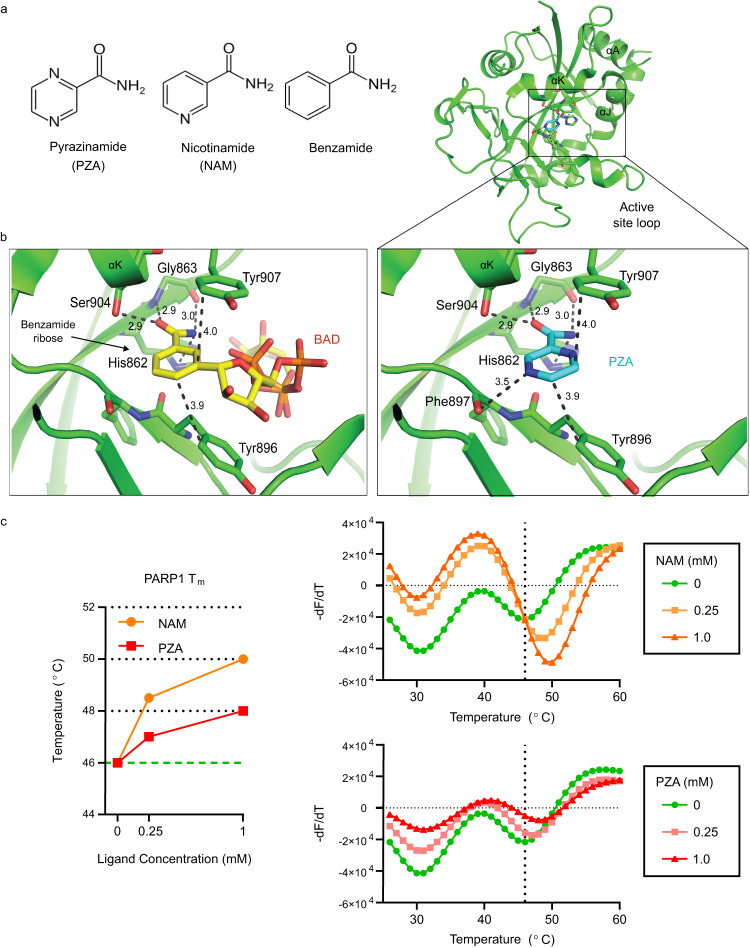
Fig. 1. PZA aligns with the PARP1 active site and shifts the PARP1 thermal stability.
a Structures of pyrazinamide (PZA), nicotinamide (NAM) and benzamide. b Cartoon of the human PARP1 catalytic domain ART fold (green) co-crystalized with the non-hydrolyzable NAD+ analog benzamide adenine dinucleotide (BAD; left) or aligned with PZA (right). Ligands and key amino acids are drawn in stick representation, and interacting residues and approximate distances (Å) are indicated. Hydroxyl groups are colored red, nitrogen atoms are blue, and the carbon backbone is yellow (BAD) or teal (PZA). Note that PZA is predicted to form one more bond with PARP1 than BAD (Phe897). The PARP1-BAD crystal structure was resolved by Langelier et al.33 and accessed from the Protein Data Bank (Accession number: 6BHV). c PZA directly binds to PARP1 and shifts the PARP1 melting temperature (Tm) in a dose-dependent manner. PARP1 Tm (left) and derivative melt curves (right) in the presence of 0.25 mM or 1.0 mM NAM (orange) or PZA (red). Derivative melt curves represent the change in SYPRO orange fluorescence intensity over increasing temperatures of PARP1 alone (green) or with increasing concentrations of NAM (top) or PZA (bottom). The temperature above 40 °C associated with the lowest point in the derivative melt curve (vertical line) was considered the PARP1 Tm (46 °C). Source data are provided as a Source Data file.