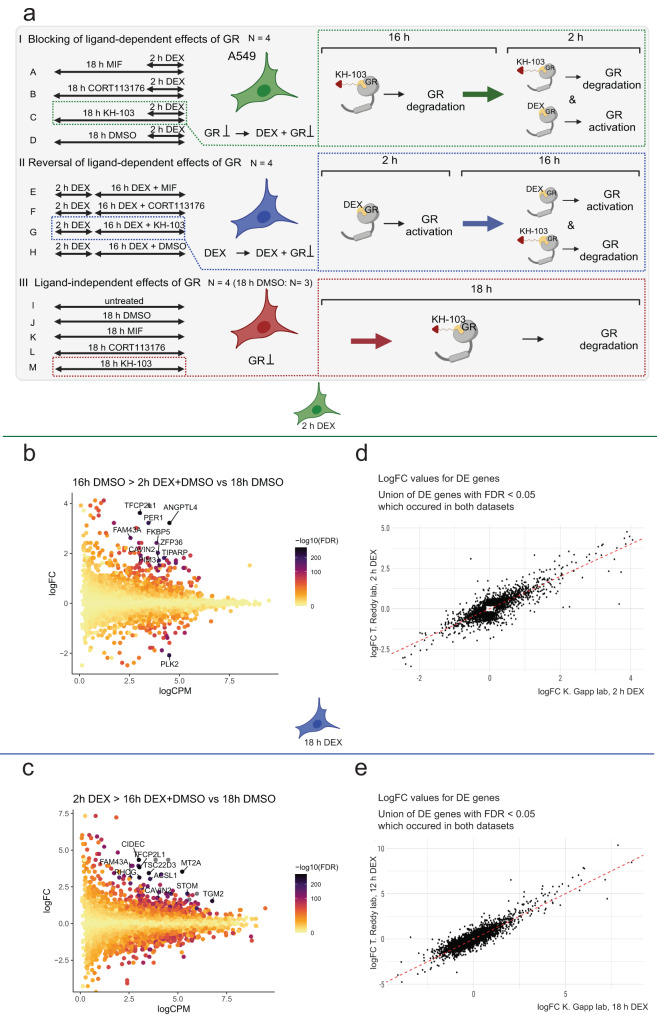
Fig. 6. The DEGs we identified upon 2 h, and 18 h of DEX treatments in A549 cells align with the other existing RNAseq and ChIP-seq datasets.
a Schematic depiction of the RNAseq experiment treatment groups in A549 cells. (n ≥ 3/group). b MA plot of 2 h DEX DEGs (K. Gapp lab). c MA plot of 18 h DEX DEGs (K. Gapp lab, current study). d Correlation of logFC values between DEGs with the false discovery rate (FDR) < 0.05 of T. Reddy lab after 2 h DEX treatment and K. Gapp lab (current study) 2 h DEX treatment. e Correlation of logFC values between DEGs with FDR < 0.05 of T. Reddy lab after 12 h of DEX treatment and K. Gapp lab (current study) 18 h DEX treatment.