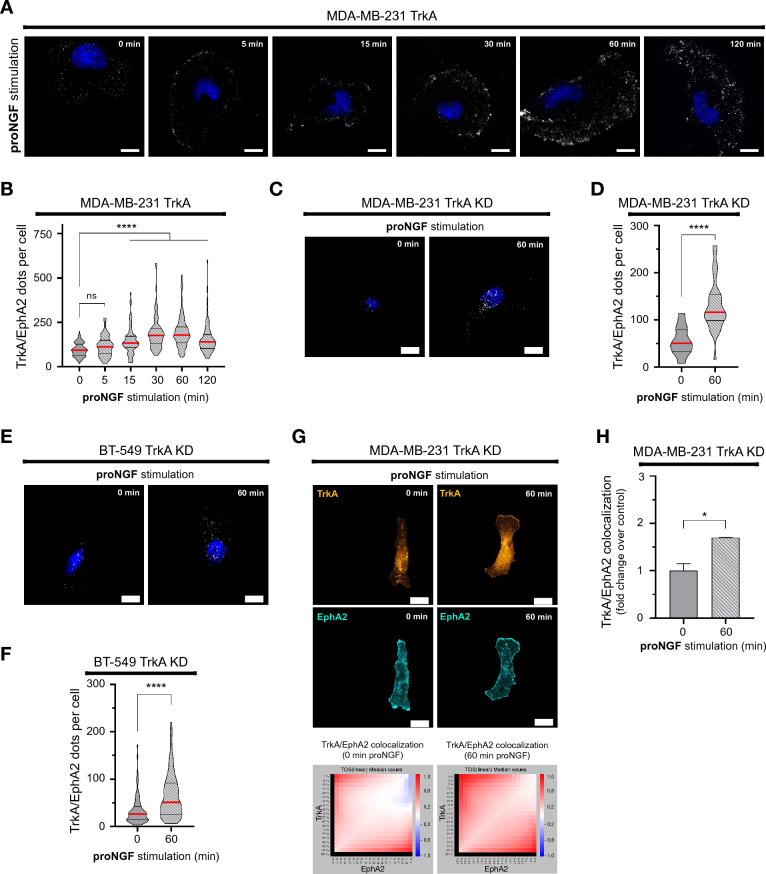
Fig. 1.
proNGF increases TrkA/EphA2 complexes in TNBC. A and B Representative overlay (A) and quantification (B) of TrkA/EphA2 PLA signal following proNGF stimulation (5, 15, 30, 60 and 120 min) in MDA-MB-231-TrkA. C–F Representative pictures of TrkA/EphA2 PLA signal following proNGF stimulation (60 min) in MDA-MB-231-TrkA-KD and BT-549 HA-TrkA KD (C & E) with respectively the associated quantification (D & F). G & H Representative confocal images of TrkA and EphA2 signal in MDA-MB-231-TrkA and TrkA/EphA2 colocalization matrix (G) with its quantification (H). Excluding experiment G and H which are performed in duplicate, data in A-F are representative of 3 independents experiments performed on 30 cells/condition each. Data in B, D and F are presented with violin plots demonstrating the median (red bold line), quartiles (dots lines), variability and density. Data in H are represented by column bar graph with SD. One-way ANOVA followed by Tukey’s test for B. Unpaired 2-tailed t test for D, F and H. *P ≤ 0.05, ****P ≤ 0.0001, ns (non-significant). For A, C and E, scale bar = 10 µm, and for G, scale bar = 15 µm