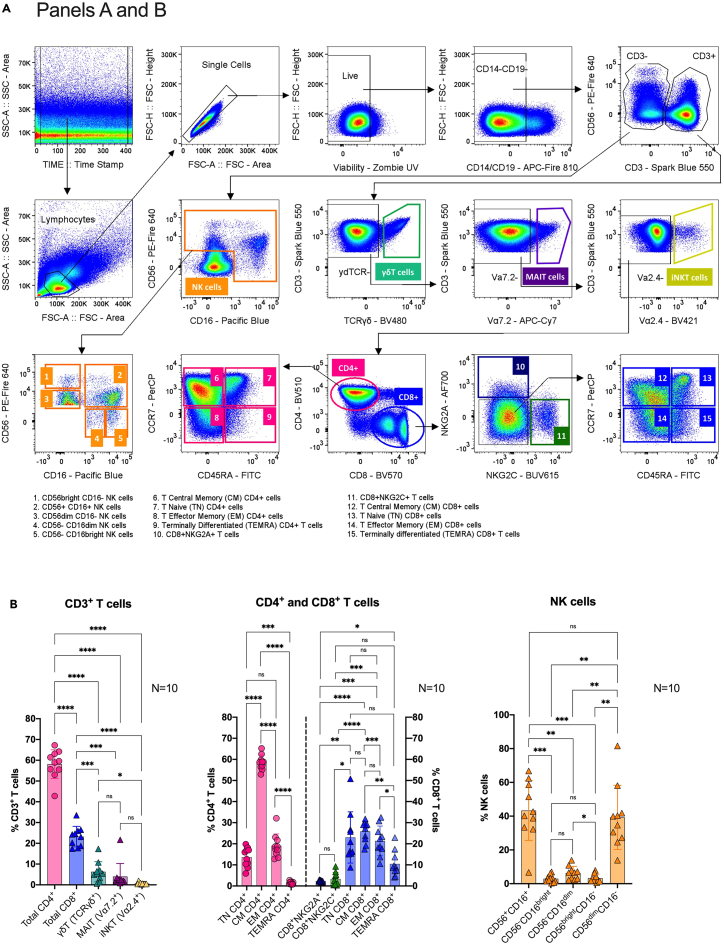
Figure 3.
Main general gating strategies for panels A and B
(A) After time lapse, lymphocyte morphology, single and live events selection, samples are gated to exclude CD14+ and CD19+ cells (monocytes and B cells). Subsequently, CD3-CD56+ cells are gated for further natural killer (NK) cells analysis using CD56 versus CD16 markers. On the other hand, CD3+ cells are gated to discriminate unconventional TCRγδ+ (γδT cells), TCRVα7.2+ (MAIT cells) and TCRVα2.4+ (iNKT cells) lymphocytes. From the remaining T cell population, cells are gated by CD4+ versus CD8+ cells. NKG2A+NKG2C- (CD8+NKG2A+) and NKG2A-NKG2C+ (CD8+NKG2C+) cells are then identified from the CD8+ T cell population. Then, the NKG2A-NKG2C- CD8+ subset as well as the total CD4+ cells are gated according to differentiation markers; CD45+CCR7+ T naïve (TN) cells, CD45-CCR7+ central memory (CM) cells, CD45RA-CCR7- effector memory (EM) cells, and CD45RA+CCR7- terminally differentiated (TEMRA) cells.
(B) Correspondingly, example graphs showing manually gated major populations (bottom figures) as proportions from their parent CD3+ T cell population (left), CD4+CD8- and CD4-CD8+ T cells (center), and NK cells (right).Bar graphs represent the means; error bars represent the standard deviation; and dots represent individual donors (N=10). One-way ANOVA with Tukey’s multiple comparison test. Significance is indicated as ∗∗∗∗P ≤ 0.0001, ∗∗∗P ≤ 0.001, ∗∗P ≤ 0.01, ∗P ≤ 0.05, or P > 0.05 (not significant (ns)).