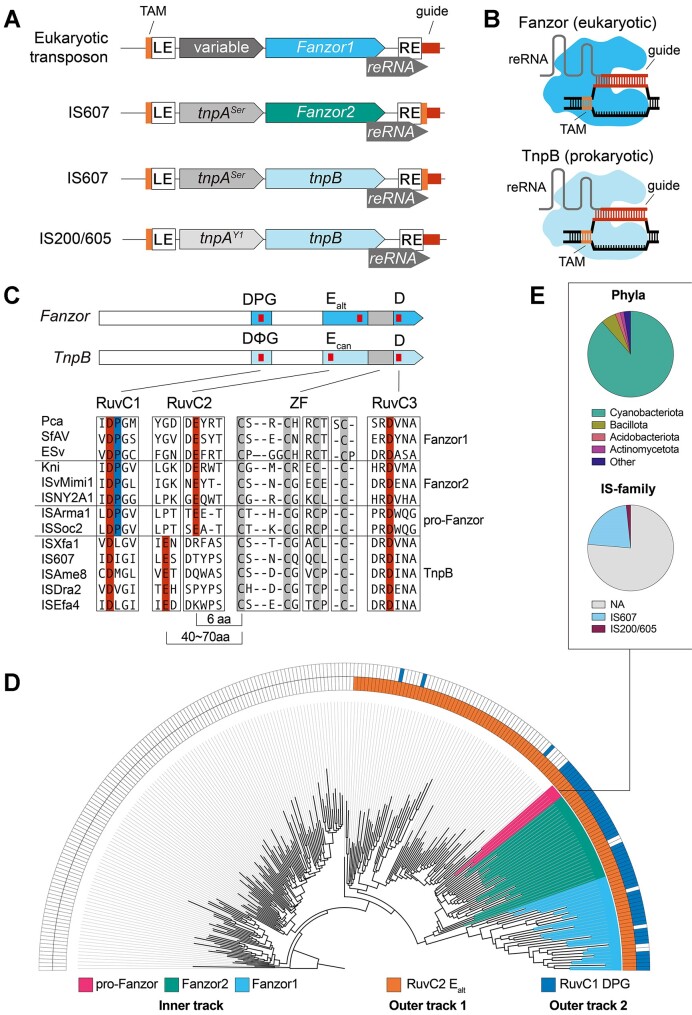
Figure 1.
Fanzors and a unique clade of TnpBs share unusual active site signatures. (A) Depiction of loci architecture of Fanzor and TnpB containing transposons. LE and RE correspond to left-end and right-end transposon boundaries. The transposon associated motif (TAM) and guide region are highlighted in orange and red, respectively. (B) Illustration of Fanzor/TnpB complexed with their right-end RNAs (reRNAs) restricting DNA. Fanzors/TnpBs recognize DNA containing both a TAM and sequence complementarity to the guide region of the reRNA. (C) Fanzors and TnpBs with RuvC (blue) and Zinc-Finger (grey) domain annotations. Red ticks indicate locations of D-E-D RuvC triad residues. Fanzors and TnpBs are differentiated by the DPG and DΦG, and Ealt and Ecan signatures in the RuvC domain, respectively. Multiple Sequence Alignment encompassing RuvC and Zinc-Finger domains of Fanzors and TnpBs is shown. (D) Maximum likelihood tree of TnpBs and Fanzors annotated by the RuvC features. See legend for the annotations depicted in the track indicated below. (E) Pie charts detailing phyla and IS family classification of the prokaryotic group of TnpBs closely related to Fanzor2s (pro-Fanzors) in (D).