Fig. 3.
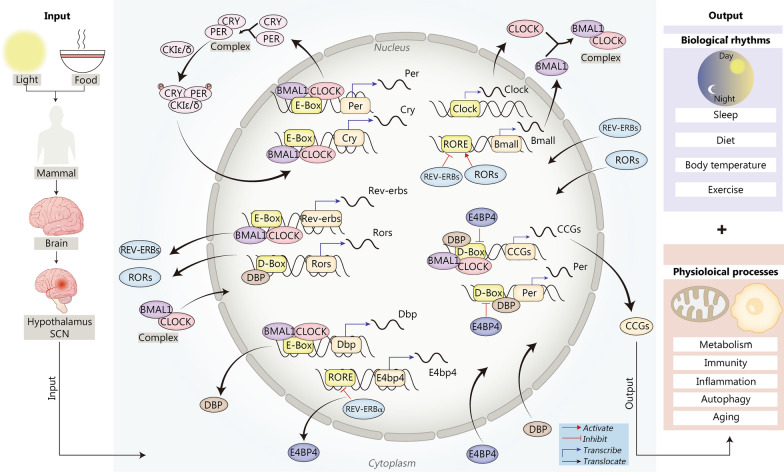
Transcription-translation feedback network of the central circadian oscillator in mammals. Input of light signal and diet signal to the SCN of the mammalian hypothalamus produces transcriptional activation of Per, which regulates the PER concentration and subsequently affects biological clock phasing. Mechanically, the core clock proteins BMAL1 and CLOCK form BMAL1-CLOCK heterodimer, translocating into the nucleus and binding to the E-box containing DNA region upstream of the promoters of downstream genes. Then, the transcription of downstream genes is activated, including Per, Cry, Rev-erbs and DBPs and other CCGs. The products of these genes partially translocate back to the nucleus, which feedback regulates the transcription of other clock genes. For example, PER and CRY form a complex that is phosphorylated by CKIε/δ, and subsequently returns to the nucleus to inhibit the activation of the BMAL1/CLOCK complex; REV-ERBs and RORs exert negative and positive regulation of Bmal1 transcription by competitively binding to the RORE in the promoter, respectively; DBP translocates to the nucleus and activates the transcription of Rors, Per, and CCGs. Subsequently, the activation of CCGs regulates the output of multiple circadian behaviors, such as biological rhythms and several physiological processes. BMAL1 brain and muscle ARNT-like 1, CCGs clock control genes, CLOCK circadian locomotor output cycles kaput, CRY cryptochrome, CKIε/δ cyclin-dependent kinase inhibitor protein ε/δ, DBP D site binding protein, PER period, ROR RAR-related orphan receptor, E4bp4 E4 promoter binding protein 4, SCN suprachiasmatic nucleus