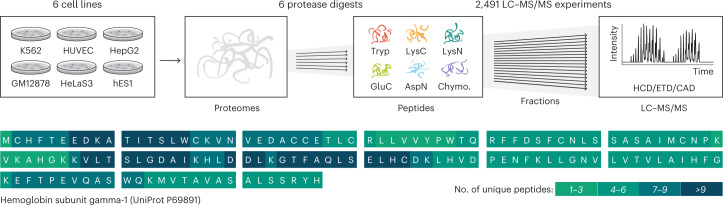
Fig. 1. Deep proteome sequencing workflow.
Six human cell lines were grown in parallel, their proteomes were isolated and then one of the six proteases was used to digest separate aliquots of each proteome in parallel. Peptides resulting from each digestion were fractionated by high-pH RP chromatography and then analyzed separately with nLC–MS/MS using HCD, ETD and CAD. The resulting data were searched with MaxQuant51,52 against the human proteome database, and over 17,000 proteins were identified by peptides that produce a median coverage of over 80%. The high coverage achieved is illustrated on the sequence of hemoglobin subunit gamma-1, with color coding to illustrate the number of unique peptides that cover each amino acid position.